ISSN: 2206-7418
Nanotheranostics 2018; 2(3):271-279. doi:10.7150/ntno.24590 This issue Cite
Research Paper
Genome-wide Transcriptional Analysis of Oxidative Stress-related Genes and Pathways Induced by CdTe aqQDs in Mice
1. Department of Toxicology and Sanitary Chemistry, School of Public Health, Capital Medical University, Beijing, 100069, China
2. Beijing Key Laboratory of Environmental Toxicology, Capital Medical University, Beijing, 100069, China
3. Xue Yuan Road Community Health Service Centers, Beijing, 100069, China
†These authors contributed equally to this work
Abstract
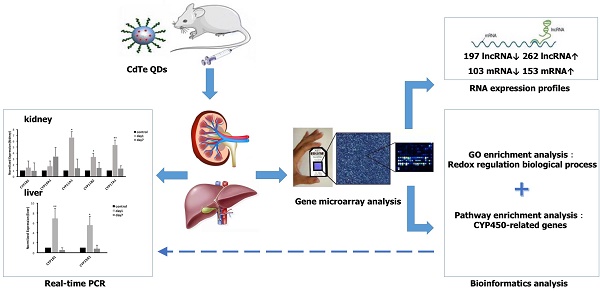
Objective: Quantum dots (QDs) has widely applied in the field of science, whose potential toxic effect has increasingly become a focus concern we need pay attention to in public health. The purpose of this article was to explore the toxicity mechanism with oxidative damage from treatment with QDs at the molecular level through a gene microarray.
Methods: Mice were administered aqueously synthesized cadmium telluride QDs (CdTe aqQDs) via intravenous tail injection of a 2 µmol/kg solution (based on the molar mass of Cd), and their kidneys were collected at 1 day in strict accordance with the programs used for treated mice. We determined the hierarchical clustering of expression ratios, enriched gene ontology (GO) terms and signaling pathways through gene microarray analysis and bioinformatics analysis in kidney tissue and screened the key enzyme genes, which were verified by real-time quantitative polymerase chain reaction (real-time qPCR).
Results: Compared to control group, 459 lncRNAs (197 down-regulated and 262 up-regulated) and 256 mRNAs (103 down-regulated and 153 up-regulated) were differentially expressed. According to biological processes in enriched GO terms, the response to a redox state played a significant role in the biological processes involved altered genes. Pathway analysis showed that the signaling pathways that involved cytochrome P450 (CYP450) enzymes had a close relationship with QDs. Among these signaling pathways, gene expression profiling revealed that selected differentially expressed mRNAs (CYP19A1, CYP1B1, CYP11A1, CYP11B2, and CYP17A1 in the kidney and CYP19A1 and CYP1B1 in the liver) were validated by real-time qPCR, resulting in expression levels of CYP11A1, CYP11B2 and CYP17A1 in the kidney and CYP19A1 and CYP1B1 in the liver that were significantly increased, however in expression levels of CYP19A1 and CYP1B1 compared with control group in the kidney, there was no significant difference.
Conclusions: Our results provide a foundation for and potential insight into the role of CYP450-related genes in QD-induced oxidative stress. QDs may produce a great deal of reactive oxygen species (ROS) by promoting high expression of CYP450 enzymes and accumulating steroid hormones, which may be an important toxicity mechanism for mediating oxidative stress and tissue damage.
Keywords: Quantum dots, Cytochrome P450 enzymes, Gene microarray, Oxidative stress

 Global reach, higher impact
Global reach, higher impact