ISSN: 2206-7418
Nanotheranostics 2017; 1(4):369-388. doi:10.7150/ntno.21136 This issue Cite
Review
Multiplexed Optical Imaging of Tumor-Directed Nanoparticles: A Review of Imaging Systems and Approaches
1. Department of Mechanical Engineering, University of Washington, Seattle, WA 98195, USA;
2. Department of Pathology, University of Washington School of Medicine, Seattle, WA 98195, USA.
Received 2017-5-22; Accepted 2017-7-8; Published 2017-8-19
Abstract
In recent decades, various classes of nanoparticles have been developed for optical imaging of cancers. Many of these nanoparticles are designed to specifically target tumor sites, and specific cancer biomarkers, to facilitate the visualization of tumors. However, one challenge for accurate detection of tumors is that the molecular profiles of most cancers vary greatly between patients as well as spatially and temporally within a single tumor mass. To overcome this challenge, certain nanoparticles and imaging systems have been developed to enable multiplexed imaging of large panels of cancer biomarkers. Multiplexed molecular imaging can potentially enable sensitive tumor detection, precise delineation of tumors during interventional procedures, and the prediction/monitoring of therapy response. In this review, we summarize recent advances in systems that have been developed for the imaging of optical nanoparticles that can be heavily multiplexed, which include surface-enhanced Raman-scattering nanoparticles (SERS NPs) and quantum dots (QDs). In addition to surveying the optical properties of these various types of nanoparticles, and the most-popular multiplexed imaging approaches that have been employed, representative preclinical and clinical imaging studies are also highlighted.
Keywords: multiplexed molecular imaging, Raman spectroscopy, hyperspectral imaging, preclinical study, tissue diagnosis, surgical guidance.
Introduction
With over 8 million deaths in 2012, cancers of all forms are one of the most prevalent and deadly diseases in the world [1]. Advanced cancer imaging methods would ideally enable the detection of cancers at their earliest stages (before metastasis) and could also enable the complete removal of tumors, which are both important for reducing mortality amongst cancer patients [2, 3]. Histopathology, which relies on the microscopic examination of physically sectioned tissue specimens, is currently the gold-standard method for cancer diagnosis. However, histopathology requires invasive tissue resection and time-consuming sample preparation, and also suffers from sampling errors. These limitations motivate alternative optical diagnostic methods.
Over the past few decades, molecular imaging has shown great potential to promote earlier and more accurate detection of cancers, as it can reveal molecular phenotypes that directly reflect the biological processes in a cancer [4-6]. While other molecular imaging modalities have been used in the clinic for cancer detection such as positron emission tomography (PET), single photon emission CT (SPECT) and magnetic resonance spectroscopy (MRS), optical imaging is attractive as it can provide images in real time with high spatial resolution and the use of non-ionizing irradiation [7, 8]. One challenge for the molecular imaging of cancers is that the molecular profiles of most cancers vary greatly between patients as well as spatially and temporally within a single tumor mass [9-12]. Therefore, to improve the sensitivity and specificity of cancer detection, multiple disease-related molecules (or biomarkers) should ideally be evaluated, thereby necessitating multiplexed molecular imaging techniques.
Unlike label-free imaging methods, which are unable to provide information about the expression of large macromolecular targets, molecular imaging using exogenous contrast agents allows one to selectively image protein biomarkers of interest with enhanced signal, image contrast, spatial resolution and speed. Various types of exogenous contrast agents can be used for multiplexed molecular imaging, such as conventional fluorescent dyes, quantum dots (QDs) and surface-enhanced Raman-scattering nanoparticles (SERS NPs). Conventional fluorescent dyes have been well-established and commercialized, in which some have been approved for use in the clinic [13]. However, in general, they have some shortcomings for multiplexed imaging. For example, their relatively large emission bandwidth of ~50 nm (FWHM) [14, 15] limits the degree of multiplexing that is possible, especially in the near-infrared region (650-800 nm) where tissue autofluorescence and water absorbance are both low [16]. In addition, multiple excitation wavelengths are often required to image multiple dyes [13]. In comparison, imaging probes such as QDs and SERS NPs have narrower emission linewidths and have the added benefit that different “flavors” of probes may be excited with a single wavelength of light, often allowing five or more molecular targets to be imaged using a single-excitation spectral-imaging system (Table 1). As will be discussed later, the use of a single excitation wavelength has major advantages for quantitative and ratiometric imaging of multiple nanoparticle probes.
In this review, after a brief introduction of some classes of nanoparticles that enable efficient and dense multiplexing, we will focus our discussion on recent advances in optical imaging systems that are designed to image these classes of multiplexed nanoparticle imaging probes (size 1-200 nm). Representative results of preclinical and clinical imaging studies are provided to highlight the value of multiplexed molecular imaging for clinical applications.
Previously reported imaging systems for multiplexed imaging of nanoparticles (DOM denotes the degree of multiplexing that was used in the study).
| Probe type | DOM | Light source | Imaging mode (system) | Signal collection (system) | Spectral resolution | Spatial resolution | Applications | Refs |
|---|---|---|---|---|---|---|---|---|
| Dye-doped silica NPs | 3 | 488 nm laser | Microscope, scanning (Olympus Fluoview 500) | 3 PMTs | 20/30 nm (bandpass filter) | Microscopic | Multiplexed monitoring of cancer cells | [23] |
| QDs | 5 | 445-490 nm, xenon lamp | Macroscope, wide-field (Maestro In-Vivo) | CCD, 500-950 nm | 10 nm (LCTF) | >25 µm | Multiplexed monitoring of lymphatic drainages | [57] |
| 5 | 330-385 nm | Microscope, wide-field (Olympus IX71) | CCD (Olympus QColor5), hyperspectral camera (Nuance), 420-720 nm | 10 nm (LCTF) | Microscopic | Single-cell molecular profiling | [58] | |
| 5 | 350 nm, mercury lamp | Microscope, wide-field (Olympus IX71) | Multispectral imaging system (Nuance), 500-800 nm | 10 nm (LCTF) | Microscopic | Multiplexed detection of rare tumor cells in tissues | [59] | |
| 5 | 405 nm laser, 25 mW | Microscope, scanning (Zeiss LSM 510 META) | META detector (32 PMT array); spectrograph (SpectroPro 150, Roper Scientific) and CCD | 10 nm (PMT) | Microscopic | Molecular profiling of cancer cells & tissue sections | [60] | |
| 3 | 445 nm, LED, 1 mW | Fiber bundle, wide-field | CCD (Retiga EXi or JAI CV-S3200N) | 20 nm (bandpass filter) | < 4 µm | Molecular imaging of cancer cells & tissues | [61] | |
| SERS NPs | 10 | 785 nm laser, 42 mW | Fiber bundle, scanning | Spectrometer (BaySpec RamanSpec RS-780), CCD (Andor DU920P-BR-DD), 830-930 nm, 1 ms - 1 s /pixel | ~1 nm | 1 mm | Multiplexed endoscopy | [52] |
| 10 | 785 nm laser, 60 mW | Microscope, scanning (Renishaw InVia) | Spectrometer, CCD, 1 s /pixel | ~1 nm | 1 mm | Multiplexed tissue imaging | [53] | |
| 5 | 785 nm laser, 15 mW | Fiber bundle, scanning | Spectrometer (Andor Holospec), CCD (Andor DU920P-BR-DD), 808-927 nm, 0.1 s /pixel | ~1 nm | 0.2-0.5 mm | Multiplexed molecular imaging of tissues | [62, 63] | |
| 4 | 785 nm laser | Microscope, wide-field | EMCCD (Andor iXon DV885), 4 wavelength channels, 5 s /channel | 4 nm (tunable filters) | 50 μm | Multiplexed molecular imaging of tissues | [64, 65] | |
| SWCNTs | 5 | 785 nm laser, 80 mW | Microscope, scanning (HORIBA-Jobin-Yvon) | CCD, 892-897 nm, 0.5 s /pixel | ~1 nm | Microscopic | Multiplexed molecular imaging of cancer cells & tissues | [55] |
Optical properties of nanoparticles for multiplexed molecular imaging
Nanoparticle imaging probes can be classified into fluorescence-based probes and scattering-based probes based on the different photophysical mechanisms that dominate. Figure 1 shows some examples of nanoparticles in each category along with their spectral profiles. Nanoparticles that are ideal for multiplexed imaging have some common optical properties, such as narrow emission peaks with symmetric Gaussian/Lorentzian profiles and a common, and often spectrally broad, excitation band. These properties facilitate the spectral unmixing of collected signals, and allow the simultaneous imaging of multiple flavors of probes.
Fluorescent probes
Fluorescent probes refer to contrast agents that absorb light of a specific wavelength and spontaneously emit light at a longer (Stokes-shifted) wavelength after a small time delay known as the fluorescence lifetime. Important examples of fluorescent probes include organic fluorescent dyes, dye-doped nanoparticles, polymer dots and QDs [13, 15, 17, 18]. While the former two examples utilize small organic fluorophores, the latter two examples are synthetic fluorophores in which semiconductor quantum states and bandgaps play analogous roles to the molecular quantum states (electronic and ro-vibrational states) that are at play in organic fluorophores.
Fluorescent dyes and dye-doped nanoparticles
Generally, fluorescent dyes and dye-doped nanoparticles have a broad emission band (50 nm or larger, FWHM, Figure 1), and often require multiple excitation wavelengths for different dyes, which are not ideal for multiplexed imaging [14, 19-21]. Note that silica nanoparticles can be loaded with multiple fluorescent dyes, which allows one to edit their emission spectra. In this way, many nanoparticle flavors can be made with different barcode emission spectra (i.e. the spectral shapes are based on the specific fluorophore combinations used). Although using multiple dyes further broadens the emission spectra, single-excitation multiplexing is possible [22, 23].
Polymer dots and quantum dots (QDs)
Polymer dots utilize semiconducting polymers that can exhibit strong fluorescence [15, 24, 25]. Some recent studies have claimed a 10-fold enhanced brightness of polymer dots over commercial fluorescent dyes and QDs [15, 26]. Polymer dots have a broad excitation range of 300 to 600 nm, enabling multiple flavors to be excited at the same wavelength [15, 27]. The first-generation polymer dots exhibited broad emission spectra that are similar to those of fluorescent dyes [15]. However, polymer dots with narrower emission spectra (36 nm to 50 nm) have been developed recently, which have made them promising candidates for multiplexed imaging [25, 26]. Some polymer dots are commercially available, such as the BD Horizon Brilliant Dyes. More details about polymer dots may be found in some recent reviews [15, 25].
Representative fluorescent probes and Raman-based nanoparticles as well as their emission spectra. The cartoons on the upper row only show one example of each probe type, and are not strictly drawn to scale. For the spectra shown on the bottom row, the dashed lines denote excitation spectra, and the solid lines denote emission/scattering spectra. The scattering spectra of the SERS NPs and carbon nanotubes are generated with 785-nm excitation. Since they utilize a scattering process, these nanoparticles can be excited across a broad range of wavelengths from the UV to NIR, but with varying efficiencies.

QDs are fluorescent semiconductor nanoparticles that most typically contain group II-VI (e.g. CdSe and CdTe), III-V (e.g. InP and InAs), IV-VI (e.g. PbTe, PbSe) or I-III-VI (e.g. CuInS2) elements [17, 28, 29]. Like polymer dots, one of the most attractive properties of QDs is their narrow and symmetric emission bands (~30 nm, Figure 1), which can be precisely tuned from the ultraviolet to infrared regions by changing the nanoparticle sizes and compositions [17, 29]. The broad absorption spectra and large Stokes shifts of QDs (Figure 1) enable simultaneous imaging of multiple flavors of QDs with single-wavelength excitation. Compared to conventional fluorescent dyes, QDs exhibit enhanced brightness and high resistance to photobleaching and chemical degradation [14, 17, 30-32]. QDs typically have a core-shell structure (e.g. CdSe core with a ZnS shell) functionalized with different coatings to optimize their quantum yields and stability [17, 28, 33]. Similar to other fluorescent probes, QDs are often coupled to a biomolecule for targeted imaging [28].
Raman-scattering-based probes
SERS NPs
Compared with signals generated via non-enhanced spontaneous Raman spectroscopy, signal enhancements of 106 to 1014 have been observed when Raman-active analytes are placed on a roughened metal surface or on metallic nanoparticles (e.g. gold or silver) [34-37]. This effect is termed surface enhanced Raman scattering (SERS). SERS was first discovered in the 1970s [38-40], and has been extensively investigated for ultrasensitive molecular detection and various biomedical applications since the late 1990s [34-37, 41-43]. A detailed explanation of the primary mechanisms that underlie the SERS phenomenon can be found in many publications [37, 44-47]. The extent of enhancement is, in part, determined by the chemical properties of the molecule [44], which means that the brightness of SERS NPs is influenced to some degree by the specific Raman reporters that are used. For example, by using Raman dyes with absorption maxima matched with NIR light sources, and a high affinity for gold surfaces, surface-enhanced resonant Raman scattering (SERRS) NPs have been developed with significantly enhanced brightness to enable ultrasensitive biomarker detection [48, 49].
SERS NPs are fabricated by attaching strong Raman-scattering molecules (i.e. Raman reporters) to the surface of noble metal cores (gold or silver), which allows the reporters to generate strong SERS signals under laser excitation (Figure 1) [46]. A protective shell stabilizes the SERS signal, preventing reporter detachment and aggregation of metallic cores. In addition, the protective shell makes the spectral output independent of the environment by preventing the adsorption of other molecules onto the metallic NP cores. In other words, the spectral fingerprints generated by the SERS NPs act as stable beacons to identify and quantify the NPs, and are completely unchanged by their chemical environment or molecular targets. A targeting molecule such as an antibody can be conjugated to the shell for targeted imaging. Details regarding the design and synthesis of SERS NPs have been discussed in some recent reviews [46, 50].
SERS NPs have several advantageous optical properties for multiplexed imaging over fluorescent dyes and quantum dots. Compared with broadband fluorescence spectra, the spectra of SERS NPs consist of a fixed combination of narrow emission peaks (1-2 nm, FWHM) [51], which allow different SERS NPs to be detected within a narrow wavelength region when excited at a common wavelength. Typically NIR light is used to minimize tissue autofluorescence and to reduce light scattering, which enables increased light penetration in tissue. Recent studies have demonstrated multiplexed imaging of 10 SERS NPs in tissues under 785-nm laser excitation [52, 53]. Similar to other nanoparticle agents like QDs, SERS NPs are photostable and do not bleach over time [54].
Other Raman-scattering-based probes
Other Raman-scattering-based probes have been investigated. For example, single-walled carbon nanotubes (SWCNTs) can have a single bright Raman peak (G band) that is tunable within a small wavelength range (1529 cm-1 to 1590 cm-1) by adjusting the ratio of C12 and C13 isotopes [55]. One study showed that five flavors of SWCNTs could be synthesized with well-separated Raman peaks (Figure 1). Multiplexed molecular imaging of cancer cells and tumor xenograft have also been demonstrated by conjugating the SWCNTs to different antibodies [55].
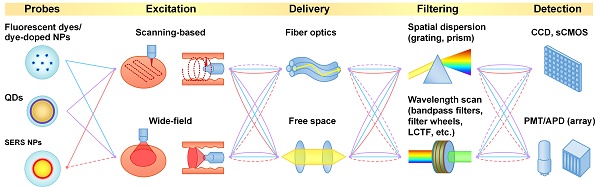
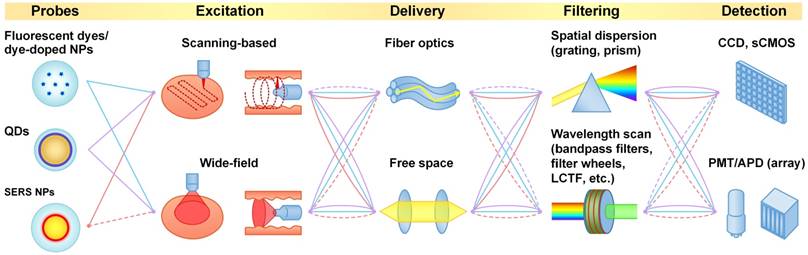
Imaging systems
In this section, we summarize representative imaging approaches and systems for the multiplexed imaging of fluorescent and Raman-based probes (Table 1).
Light sources
Nanoparticle imaging probes can be excited with a variety of light sources such as lasers, light-emitting-diodes (LED) and high-intensity discharge lamps (e.g. mercury and xenon). For single-wavelength excitation applications (the focus of this review), lasers are often used due to their high stability [56], their superior brightness (high directionality and radiant flux, with low etendue), and their narrow linewidths. The narrow linewidth is essential for generating narrow Raman-scattering spectra and is also preferred for fluorescence imaging because wide-band excitation can broaden the spectra of the tissue autofluorescence background. The excitation wavelength is constrained by the optical properties of the imaging probes. For example, fluorescent dyes and dye-doped nanoparticles typically have an optimal excitation wavelength that is close to their emission peak due to their relatively short Stokes shift (typically 20-50 nm), so the collection filters need to be carefully selected to reject excitation light while collecting as much fluorescence signal as possible. UV and blue excitation is often used to excite QDs due to the increased absorption (extinction coefficient) of QDs at UV wavelengths. Red or NIR excitation is often used to image Raman-scattering-based probes to minimize tissue autofluorescence, with the added benefit that the Stokes shift of Raman scattering can be considerably higher than for fluorescence (i.e. better separation of illumination and scattered light).
Signal collection
Signal collection methods differ amongst various imaging probes. For fluorescent probes that have a relatively wide emission spectrum (e.g. fluorescent dyes, dye-doped nanoparticles and polymer dots), bandpass filters with a bandwidth of 20-30 nm are often used to separate the signals from different fluorophores. When 3 or more fluorescent probes are multiplexed, spectral overlap is often unavoidable and light collected in individual wavelength channels comes from multiple fluorophores (Figure 2). In that case, linear unmixing of the collected signals is often used [66, 67]. While filter-based imaging methods typically collect signals from a limited number of wavelength channels, hyperspectral and spectrometer-based imaging devices have been developed to disperse signals into many more wavelength channels (up to thousands), which can reveal subtle spectral features and offer more accurate signal separation (linear unmixing [68-70]). In this review, we use the terms “multispectral,” “hyperspectral,” and “spectral imaging” to refer to imaging modalities that acquire two-dimensional images across a range of visible and infrared wavelengths. Hyperspectral imaging generally covers more spectral bands (up to a few hundred) with higher spectral resolution than multispectral imaging (e.g. RGB color cameras). Spectral imaging with moderate spectral resolution (5-20 nm) is often used for the detection of fluorescent dyes and QDs, and spectral imaging with higher resolution (~1 nm) is often used for detecting SERS NPs.
Schematic of the linear unmixing of signals from multiple fluorophores. The colored circles represent beads labeled with three different fluorophores. The emission spectra of the fluorophores overlap and cannot be well separated with bandpass filters. However, by using a linear unmixing algorithm based on the known signal contributions (the reference spectra) of each fluorophore in each detection channel, the overlapped signals can be clearly separated. The composite image is color-coded based on fluorophore type/flavor.
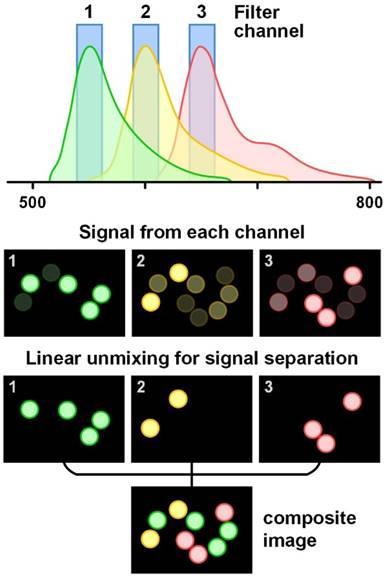
Signal unmixing
Signal unmixing is a critical aspect of spectral-imaging approaches. When multiple imaging probes are imaged simultaneously, accurate separation of signals from the different probes is essential for quantitative analysis of the molecular targets (Figure 2). Linear unmixing methods are often used [71], which have been shown to enable accurate separation of multiplexed probes visualized with both bandpass-filter-based imaging (multiple discrete channels) [72] or hyper-/multi-spectral imaging (many sequential channels) [66]. Linear unmixing (e.g. direct classical least squares) assumes that each measured spectrum is a linear combination of a known basis set of reference spectra, which often includes both the reference spectra of the imaging probes as well as tissue autofluorescence and other background components (if known). Linear unmixing methods have been broadly used for multiplexed imaging of fluorescent dyes, QDs and SERS NPs, as discussed in many publications [66, 70, 73-76]. For QDs or SERS NPs that can be multiplexed with single-wavelength excitation, photobleaching is not a concern, which simplifies the linear unmixing algorithms. In contrast, multiplexed imaging of fluorescent dyes often requires more complex algorithms due to the involvement of multiple excitation wavelengths as well as photobleaching issues. Although the spectral shape of fluorescent probes do not change with excitation wavelength, their emission intensities do change [69]. Therefore, for each fluorophore, multiple reference spectra are often acquired for the linear unmixing process to accurately represent them under different excitation conditions. With 6 excitation wavelengths, multiplexed imaging of 16 fluorescent dyes (or 120 combinations of two of the 16 dyes) has been reported, using either a Nikon A1 or a Zeiss 710 laser scanning confocal microscope (32 channels, bandwidth <10 nm per channel) [69]. The excitation power of the 6 lasers must be kept the same for reference acquisition and multiplexed imaging [69]. In addition, photobleaching must often be considered [77].
Tissue background spectra are often included in the linear unmixing process when the background signals are not negligible compared to the probe signals. In practice, variations in tissue background are often observed due to heterogeneous tissue components. For example, tumor, muscle and fat exhibit different autofluorescence spectra [78]. Variations in hemoglobin concentration in muscle, or variations in collagen, NADH, and/or melanin in skin, can also alter tissue background spectra [79]. Many methods have been proposed to increase the robustness of the unmixing algorithms to tissue background variations, such as adding a low-order polynomial term to compensate for slight background changes [80], allowing the baseline to vary [81-83], allowing the peaks of the reference spectra to vary [84], and incorporating the principal components of variable background spectra as reference spectra [70]. Allowing the background spectra to vary according to a low-order polynomial model can effectively account for minor broadband background variations without acquiring numerous background spectra [80]. Principal component analysis (PCA) requires additional background measurements (training sets) from many potential sources of tissue background (tumor, normal, fat, skin, etc.) to extract a few principal component spectra, which can be used as reference spectra to account for moderate or high background variations [70, 75]. The combination of PCA and a polynomial model into a linear-unmixing algorithm has been shown to be a robust method to deconvolve signals of multiplexed SERS NPs in heterogenous tissues [62, 85](1).
(1)
where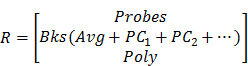
S = measured spectral data
wn = weight of spectral component n
Rn = reference spectrum of component n.
The reference spectra include contributions from the imaging probes (Probes), tissue background and other background components (e.g. tissue substrate, buffer). If a background varies (e.g. heterogenous tissue), the average spectrum (Avg) and the first 2 or 3 principal component spectra (PC1, 2…) of numerous prior acquired background spectra may be used as reference spectra. In addition, a low-order polynomial term (Poly) may be added to account for unknown broadband variations.
Imaging approaches
Scanning-based imaging
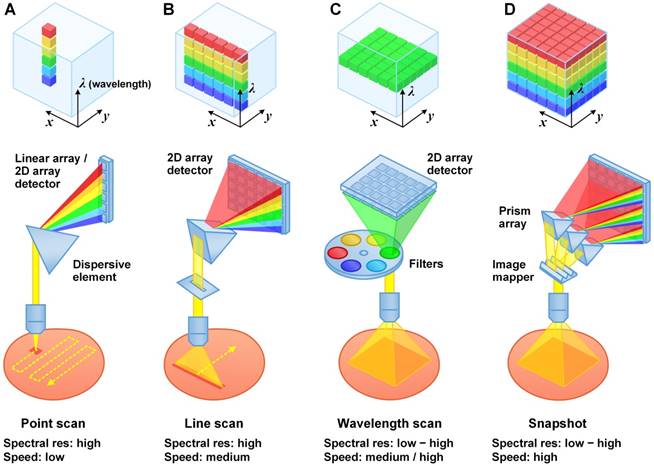
The two major spectral imaging categories are scanning-based imaging and wide-field imaging (Figure 3). The former is usually implemented by scanning a collimated or focused laser beam (in the form of a spot or a line) across the specimen (Figure 4A and 4B). For example, raster scanning may be achieved by steering the laser beam with a galvanometric scanning mirror (high speed) or translating the specimen with a mechanical stage (low speed) [86]. Rotational scanning is often utilized in endoscopy to image hollow organs such as the esophagus and colon [87, 88]. Compared with wide-field imaging approaches, scanning approaches can provide improved spectral resolution by collecting a highly resolved spectrum at each point or line of pixels on the sample, but at the cost of decreased imaging speed. High spectral resolution enables accurate demultiplexing and quantification of imaging probes.
A variety of detectors can be used for scanning-based imaging, such as photomultiplier tubes (PMT), avalanche photodiodes (APD), charged coupled devices (CCD), intensified CCDs (ICCD), electron-multiplying CCDs (EMCCD), and scientific complementary metal-oxide semiconductor (sCMOS) arrays. Among them, the PMT is the most sensitive detector but suffers from a low dynamic range at any given gain setting. In addition, high costs prohibit the manufacturing of high-pixel-count PMT arrays (low-pixel arrays such as 1×32 and 8×8 arrays are available at reasonable costs). Furthermore, most PMTs are only sensitive to light at 200 - 900 nm (common photocathode materials lose 99% quantum efficiency above 800 or 900 nm). Nevertheless, PMTs are often used in scanning-based imaging approaches to achieve high imaging speed. Laser scanning confocal microscopes equipped with 32-channel PMT arrays are commercially available for hyperspectral imaging of multiplexed fluorescent dyes and QDs (e.g. Zeiss LSM 710/780, Nikon A1, ~10 nm spectral bandwidth per channel). APDs, though slightly less sensitive than PMTs, are much less expensive and exhibit superior dynamic range. A recent study even showed that an APD can achieve better sensitivity than a PMT in the NIR range (650-1000 nm) [89]. CCDs are often used for spectral imaging. A full spectrum can be recorded by dispersing the collected light along one dimension of the CCD through the use of a prism or grating. The large number of pixels in CCD arrays can be utilized for flexible tuning of spectral resolution by binning adjacent pixels (in the spectral direction). Spectral resolutions of 0.5-2 nm are often utilized for multiplexed imaging of Raman-scattering-based probes (e.g. SERS NPs and SWCNTs) [48, 52, 55, 90, 91]. However, our recent study showed that a much lower spectral resolution of 4-8 nm can still allow for accurate unmixing of up to 5 SERS NP flavors, suggesting the potential of using highly sensitive detector arrays (e.g. 16/32-channel PMT/APD array) as an alternative to enhance imaging speeds [92] (Figure 5).
Schematic of optical imaging approaches for different imaging probes. The solid lines represent widely used approaches, and the dash lines represent rarely used or potential approaches.
Typical (hyper)spectral imaging approaches. (A) Point scan. (B) Line scan (i.e. “pushbroom”). (C) Wavelength scan. (D) Snapshot.
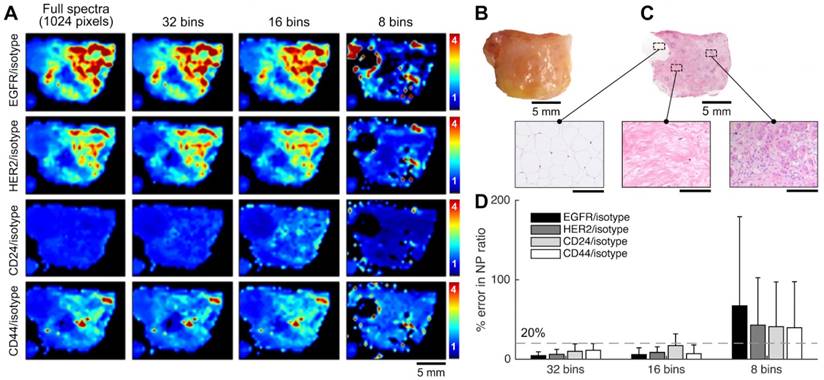
Raman-enabled molecular imaging of a human breast tissue specimen stained with a 5-flavor SERS NP mixture (EGFR-, HER2-, CD24-, CD44-, and isotype-NPs, 150 pM/flavor) - a comparison of spectral imaging and unmixing using 1024 and binned (32, 16 and 8) spectral channels. The mixture of SERS NPs was topically applied on fresh tissue surfaces for 5 min, followed by a 10-s rinse-removal step in PBS and raster-scanned spectral imaging with a 785-nm laser (3 min). (A) Ratiometric images of a human breast tissue specimen. From top to bottom, the rows display ratiometric images of EGFR/isotype-NP, HER2/isotype-NP, CD24/isotype-NP and CD44/isotype-NP. From left to right, the columns display ratiometric images obtained with a decreasing number of spectral channels. (B) A photograph of the tissue specimen. (C) H&E histology of the specimen, with higher magnification views of fat (left), normal breast tissue (middle), and tumor (right). Unlabeled scale bars represent 200 μm. (D) Average error (%) in the measured NP ratios when using spectral compression in comparison to the gold-standard images (1024 spectral channels). The error bars represent the standard deviation amongst all pixels in the image. These results showed that a low spectral resolution of 4 nm (32 bins) or 8 nm (16 bins) may still allow accurate unmixing of 5 SERS NP flavors, suggesting the potential of using advanced detector arrays (e.g. 16/32-channel PMT/APD array) to achieve high imaging speed. Reproduced with permission from [92].
Wide-field imaging
Here, we define “wide-field imaging” as a method that illuminates a region and detects the emitted light from an array of points (pixels) within the area using a camera (a 2D detector array). Compared with scanning-based imaging methods, wide-field imaging eliminates the need for mechanical scanning and can image a large area with one acquisition (Figure 4C and 4D), which is typically simpler and faster. Wide-field imaging methods are often utilized to achieve high spatial resolution, including super-resolution with structured illumination [93]. Since wide-field imaging typically cannot reject out-of-focus light efficiently, samples with micron-scale thicknesses (e.g. cells or tissue sections) are typically necessary to obtain high-contrast images. However, there are exceptions when certain imaging probes are used. For example, large nanoparticles (>100 nm) topically applied on thick tissues have a negligible penetration depth of 10-20 µm [62], which ensures that the nanoparticle signals originate from a thin layer on the tissue surface, eliminating the need to physically section the tissue. Another strategy is to limit light penetration, by either using deep UV excitation, which ensures that only the fluorescent dyes near the tissue surface are excited [94], or using “pseudo optical-sectioning” approaches such as structured illumination microscopy (different from SIM for super-resolution) to remove some of the out-of-focus background [95-98].
CCDs and newly developed scientific CMOS (sCMOS) sensor arrays are the most widely used detector arrays for wide-field imaging, allowing for the acquisition of millions of spatial pixels with one exposure in time. Rapid hyperspectral wide-field imaging can be achieved by either acquiring two-dimensional (2D) images at multiple wavelength channels over time (Figure 4C) [65] or taking a snapshot to acquire both spatial and spectral information (sometimes referred to “snapshot hyperspectral imaging”) (Figure 4D) [99-101]. The former approach can achieve a high spectral resolution (channel bandwidth) of 1-5 nm by using monochromators, filter wheels, or electronic tunable filters such as liquid crystal tunable filters (LCTFs) and acousto-optic tunable filters (AOTFs) [100]. Although expensive, LCTFs and AOTFs are preferred for hyperspectral imaging since they are faster, more compact and stable, and offer increased spectral selectivity than filter wheels [102, 103]. The main disadvantage of filter-based spectral imaging is low light throughput and prolonged imaging times due to the fact that only one filter channel is collected at a time and the rest of the photons are discarded. In comparison, the latter “snapshot” approach typically utilizes a combination of image-division elements and dispersive elements (typically a prism or diffractive optical element) to image a 2D field onto sub-regions of a detector array such that all spatial pixels and spectral channels are imaged simultaneously (Figure 4D shows the image mapping spectrometry approach [104, 105]; detailed discussion of all snapshot imaging approaches can be found in [101, 106]). Snapshot hyperspectral imaging can enable enhanced light throughput and imaging speed, but may require a tradeoff between spatial and spectral resolution since a limited number of camera pixels must be used to collect signals from both different spectral channels as well as different spatial positions [100, 107, 108]. Additional details regarding multispectral and hyperspectral imaging systems can be found in some recent reviews [100, 101, 107]. Both filter-based and snapshot approaches have been extensively utilized for wide-field imaging of multiplexed fluorescent probes [103, 107]. For Raman-scattering-based probes, the slow scanning approach is still preferred, due to a desire for higher spectral resolution (more spectral channels). However, wide-field imaging is also applicable. For example, one group demonstrated wide-field imaging of 4 flavors of SERS NPs in mouse tissue using a tunable filter (4 channels with 4-nm bandwidth per channel), which enabled the imaging of a large field of view of 2.2 cm2 at 50-µm resolution [64, 65]. Further work is needed to explore the snapshot approach, which is theoretically the most efficient way to achieve hyperspectral imaging when a balance between spectral and spatial resolution is desired. For example, as discussed in Section 3.4.1, a spectral resolution of 4-8 nm may be sufficient to allow a snapshot imaging system to image and demultiplex 5 or more SERS NPs simultaneously ([92], Figure 5).
Fiber optic delivery
Over the past few decades, the development of fiber optics technologies has greatly broadened the applicability of optical imaging for in vivo clinical and preclinical applications such as endoscopy, point-of-care imaging/sensing with portable devices, and imaging of live animals [52, 109-115]. Fiber optic delivery can be used in both scanning-based and wide-field imaging approaches. Representative fiber optics-based scanning methods have been summarized in other review articles [110, 111]. Some examples of scanning methods used in multiplexed molecular endoscopy are provided in Section 4.1 (Figure 6 and 7).
Combining wide-field spectral imaging with fiber optics can enable the development of versatile tools for rapid clinical diagnosis. For example, bandpass-filter-based wide-field imaging of 3 QDs has been demonstrated with a 1-mm-diameter fiber bundle that contains 30,000 fibers [61]. Fiber bundles have also been adopted for snapshot hyperspectral imaging, by offering the flexibility to spatially rearrange image pixels [101, 106]. A simple strategy is to rearrange a 2D-array image into a 1D-array image so that the collected light can be dispersed onto a 2D detector in a way that facilitates image processing [116-119]. This snapshot method can achieve high spectral resolution. One study reported real-time snapshot imaging with a fiber bundle to acquire 44×40 spatial pixels × 300 spectral channels (400 to 1100 nm, ~2.4 nm spectral resolution) [119]. More sophisticated image-mapping methods can be used when high spectral resolution is not desired. For example, a snapshot hyperspectral imaging endoscope was developed with an image mapper to break an image into 24 subimages. This endoscope could acquire datacubes (350×350 pixels×48 channels, spatial resolution 100 µm) at a speed of 5.2 cubes/s for reflectance and fluorescence imaging [105]. Another recent study presented a multimodal endomicroscope that combined high-resolution confocal fluorescence imaging with wide-field snapshot multispectral imaging [120]. Ex vivo imaging of human oral tissue specimens demonstrated the capability of the endomicroscope to rapidly localize abnormal lesions within a 5-mm field of view followed by high-resolution multispectral imaging to determine the presence and degree of neoplasia [120]. These techniques can potentially be used to enable surgical guidance, early cancer detection, and molecular profiling/staging of diseases. An extended discussion of these types of clinical applications and related imaging systems will be provided in the following section.
Some limitations of fiber bundles include: undersampling due to limited numbers of fibers in flexible bundles, reduced spatial resolution and light throughput due to the space between fiber cores, and missing image pixels due to fiber damage (almost unavoidable when hundreds/thousands of fibers are used, as shown in [118]).
Cancer imaging with multiplexed imaging probes
Spectral-imaging techniques have been increasingly used to improve the detection of cancers, including those of the cervix, breast, gastrointestinal tract, skin, prostate, trachea, oral cavity, brain and lymph nodes [100, 121]. These approaches have either analyzed the intrinsic optical characteristics (absorption, scattering, and fluorescence) of tissues or have visualized the aberrant expression of cancer biomarkers with targeted exogenous imaging probes. In this section, we will focus on the latter, where representative systems used for various clinical applications will be introduced.
Early detection of gastrointestinal cancer
Gastrointestinal (GI) cancers, including those of the esophagus, stomach, pancreas, liver and colon, are some of the most prevalent diseases worldwide [122]. Early detection is key to patient survival, and could be aided by molecular imaging technologies. However, accurate detection is hampered by tumor heterogeneity - the variability in molecular expression patterns exhibited between patients and within patients over time, which motivates the imaging of multiple biomarkers. Although very few nanoparticle reagents have been approved for human use [123, 124], studies with animal models have demonstrated the feasibility of multiplexed molecular endoscopy in vivo with fluorescent probes and SERS NPs.
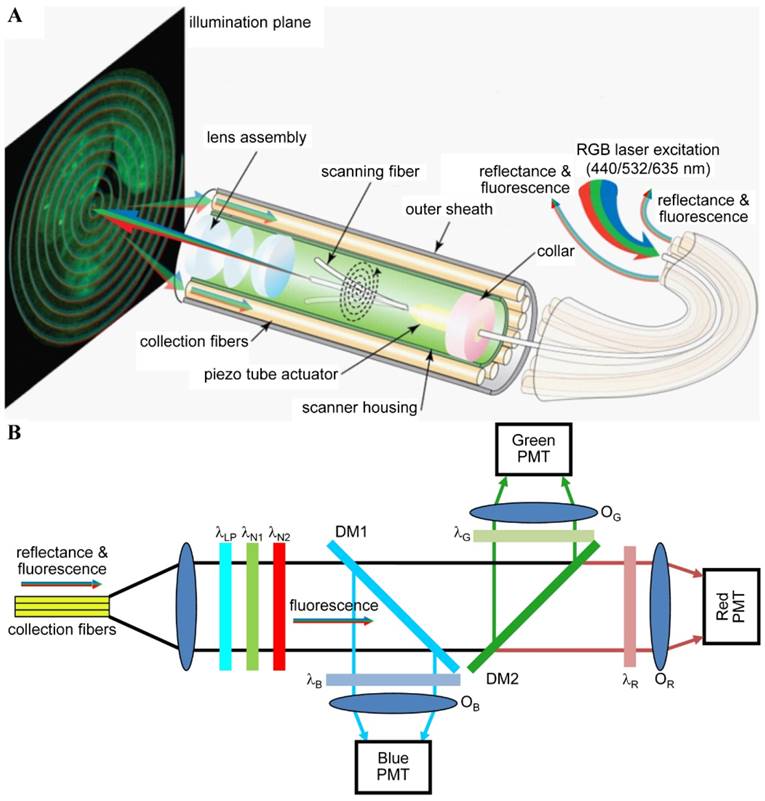
Multispectral scanning fiber endoscope. (A) Optical design. RGB laser excitation (440, 532, and 635 nm) is delivered into a single-mode optical fiber that is scanned in a spiral pattern by a piezo tube actuator and focused onto the tissue (illumination plane) by a lens assembly. Fluorescence is collected by a ring of 12 collection fibers mounted around the periphery of the scanner housing, protected by an outer sheath. (B) Fluorescence detection. Reflectance from RGB laser excitation is removed using a combination of longpass (λLP = 450 nm) and notch (λN1 = 532 nm and λN2 = 632.8 nm) filters. Fluorescence is deflected into individual RGB channels using dichroic mirrors DM1 (λC = 460 nm) and DM2 (λC = 550 nm) and an additive dichroic filter set (λR, λG, and λB) prior to detection with PMTs. Reproduced with permission from [125].
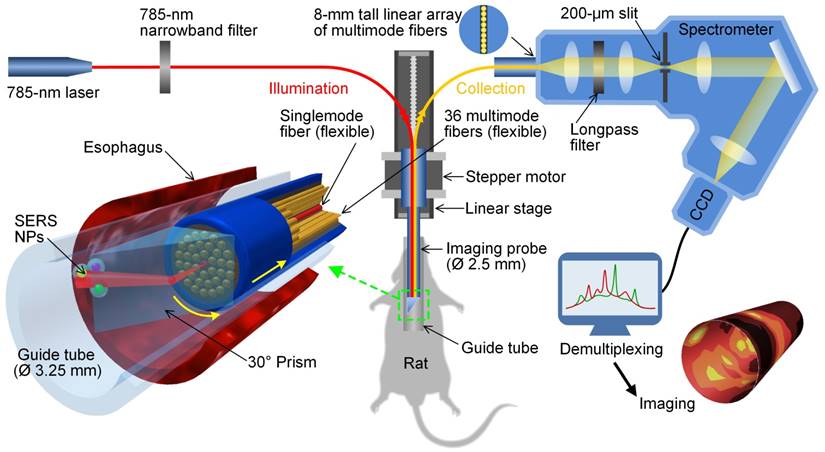
Schematic of a customized spectral-imaging endoscope to detect multiplexed SERS NPs within the esophagus of a rat. The fiber-bundle imaging probe rotates between ± 180° as it is slowly pulled out of the esophagus. The inset on the bottom left is a zoom-in rendering of the prism and fiber-bundle imaging probe (distal end) within a glass guide tube and rat esophagus. Reproduced with permission from [126].
Simultaneous imaging of three fluorophores in a mouse colon has been demonstrated with a multispectral scanning fiber endoscope with a 1.2-mm outer diameter (OD) [87, 112, 125]. Three laser sources (440, 532 and 635 nm, <2 mW/each) were delivered simultaneously through a single-mode fiber within the distal tip that was scanned in a spiral pattern (Figure 6A). Fluorescence signals were collected by a ring of 12 optical fibers (not scanned), dispersed into 3 spectral channels using dichroic beam splitters, and focused onto separate PMTs for detection (Figure 6B). The 3 fluorescent dyes were chosen with minimal spectral overlap so that signal unmixing was not necessary. The endoscope can potentially be used for the early detection of colorectal cancer via real time in vivo molecular imaging of multiplexed targeted agents.
Several spectral imaging endoscopes have been developed for multiplexed imaging of topically applied SERS NPs (up to 10 flavors) in a rat esophagus or the colon of swine [52, 113, 126, 127]. A common feature of these spectral endoscopes has been the use of a fiber bundle with a centrally located single-mode or multimode illumination fiber (785 nm) that is surrounded by many multimode fibers for light collection (Figure 7). Since a point-scanning approach was used, the illumination spot size at the tissue surface (0.5 or 1 mm) defined the spatial resolution of these systems. While the multimode collection fibers were arranged in a compact circular bundle at the distal tip of the imaging device, the fibers were rearranged into a linear array at the proximal end, which served as the entrance slit of a spectrometer (a configuration that has also been used for acquiring intrinsic Raman spectra from tissues [128]). Within the spectrometers used in these detection systems, collected light is dispersed by a grating onto a CCD array, where full vertical binning (FVB) is used to sum the contribution from all of the collection fibers to generate a spectrum. Rotational scanning is achieved by attaching a prism [126] or some other motorized component [127] to the tip of the fiber-bundle probe. Note that a topical application approach was used in these studies, which enabled rapid binding of SERS NPs to the cell-surface biomarkers on the lumen of GI tract (in <10 min [113]) with minimal systemic uptake [129-131]). Using topically applied multiplexed SERS NPs, these endoscopes can quantify multiple biomarkers over large areas of the esophagus or colon, in a time period that is consistent with routine endoscopy procedures, thereby offering the potential for early detection of GI cancers.
Surgical guidance
Surgery remains an effective treatment strategy for most types of solid tumors. However, high local recurrence rates occur in many cases due to incomplete tumor resections (i.e. positive margins). Examples include breast conserving surgery (20%-50% positive margin rates) [132], radical prostatectomy (11%-38% positive margin rates) [133] and head and neck tumor resections (11-16% positive margin rates) [134-136]. Molecular imaging has been explored to visualize tumors during surgery with the goal of maximizing tumor resection and preservation of important normal structures, including cosmetically sensitive tissues. In most cases, improved tumor resection is correlated with improved patient outcomes, and being able to successfully resect the tumor in one operation is important for minimizing patient trauma (physically and emotionally) and decreases overall health-care costs. Fluorescence-guided surgery has been demonstrated in clinical trials for decades using approved fluorescent dyes (e.g. indocyanine green and methylene blue) and commercialized imaging systems [137, 138]. However, a major hurdle for molecular image-guided surgery is the molecular heterogeneity of tumors, which can potentially be overcome by imaging multiple biomarkers. The following paragraphs describe a few examples of such approaches under development.
Simultaneous imaging of 5 QDs in mice has been demonstrated to monitor lymphatic drainage, which can potentially predict the route of cancer metastasis and the resection of involved nodes during surgery [57]. The imaging was carried out using a Maestro In-Vivo Imaging System (CRI Inc.), which was used to excite 5 QDs at 445 - 490 nm with a xenon lamp. QD fluorescence was collected in the range of 500 - 950 nm at 10-nm increments through the use of a LCTF (at exposure times of several seconds per wavelength channel). Regions of interest from a mouse were imaged with this system in a few minutes.
High grade gliomas (HGG), in particular glioblastoma multiforme (GBM, grade IV astrocytoma), are aggressive brain tumors that result in poor survival (patients die on average 12-15 months after diagnosis) [139]. Although surgical resection of GBM tumors has shown efficacy for improving patient survival [140, 141], complete tumor resection is challenging due to the inability to image infiltrating tumor cells intraoperatively with microscopic resolution and high specificity. Several fluorescence guidance technologies have been developed to maximize the extent of resections, in which a single fluorescence contrast agent was imaged [142, 143]. Other studies have explored the use of nanoparticles for guiding brain tumor resection, such as QDs [144-148] and SERRS NPs [149-151], both of which allow for multiplexed imaging. One study demonstrated simultaneous imaging of two SERRS NPs that were systemically administered in a GBM mouse model (Figure 8), in which a non-targeted SERRS NPs enabled delineation of the main tumor through the enhanced permeability and retention (EPR) effect, and a second integrin-targeted SERRS NPs enabled more-accurate visualization of the extent and the diffuse margins of the main tumor (including isolated distant tumor cell clusters of less than 5 cells) [151]. In this study, fixed brain sections (1-mm thick) were imaged instead of living mice to better correlate the Raman images with histological images (Figure 8D and 8E). Imaging was performed with a Renishaw inVia Raman microscope with 785-nm laser excitation (10-100 mW), a 5× or 20× objective (Leica), and the StreamLine line-scanning mode. The Raman spectra were unmixed using a direct classical least squares (DCLS) algorithm that is embedded in the Wire 3.4 Raman imaging software (Renishaw) to generate images of the two SERRS NPs.
Approximately 200,000 patients are diagnosed with early-stage breast cancer each year in the United States, for which breast-conserving surgery (a.k.a. lumpectomy) is a standard intervention [152]. Unfortunately, 20% - 50% of these patients require additional surgery if post-operative pathology reveals positive margins [132]. A wide variety of intraoperative guidance techniques have been developed to identify residual tumors at the surgical margins [153], with varying degrees of success and limitations. Breast cancer is a well-studied disease with established biomarkers and molecular subtypes, which are commonly evaluated by pathologists to aid diagnosis and to guide treatment decisions. Therefore, molecular imaging can potentially enable highly sensitive and specific detection of residual tumors, through quantification of multiple biomarkers intraoperatively. Early in 2007, multiplexed molecular imaging of 5 biomarkers in formalin-fixed paraffin-embedded (FFPE) tissue sections was demonstrated with 5 QDs, showing the feasibility to identify breast tumors with heterogenous molecular profiles [60]. Recently, a SERS NP-based intraoperative imaging technique has been developed to rapidly quantify multiple cancer biomarkers at the margins of freshly resected breast tissues [62, 63, 154]. This technique circumvents toxicity and sterility concerns by staining and imaging fresh surgical specimens ex vivo, and mitigates a number of misleading nonspecific effects through ratiometric imaging of targeted versus untargeted NPs to achieve unambiguous biomarker quantification [154-156]. After 5 min of staining, 10 s of rinsing and < 5 min of raster-scanned imaging of tissue specimens, 4 biomarkers were simultaneously quantified across the entire surgical margin, which enabled this technique to achieve 89.3% sensitivity and 92.1% specificity for the detection of breast carcinoma [63]. The entire process requires <15 min in total, which is acceptable for time-constrained intraoperative conditions [62] (Figure 9). The NP-stained tissue surfaces were imaged with 785-nm excitation (<20 mW) using a customized spectral-imaging system (similar to the spectral endoscope described in Section 4.1 [126]), which achieved a raster-scanned imaging speed of >3 cm2/min (integration time: 20-50 ms; pixel spacing: 0.5 mm).
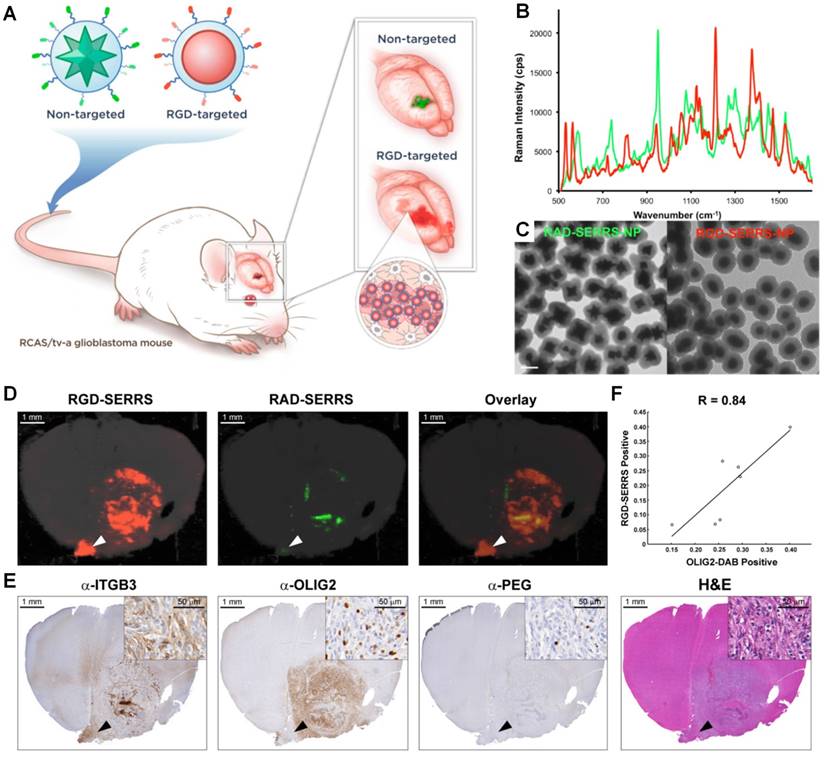
Integrin-targeted SERRS nanoparticles enable the detection of bulk and infiltrative glioblastomas. (A) Conceptual figure outlining integrin-based detection and delineation of glioblastoma with cRGDyK-conjugated SERRS nanoparticles. Due to the enhanced permeability and retention (EPR) effect of the GBMs, accumulation of the non-targeted RAD-SERRS nanoparticles (green) was limited to the bulk tumor, while the RGD-SERRS probe (red) could be detected in both the bulk tumor and the infiltrating tumor cells due to targeting of overexpressed integrin receptors. (B) SERRS spectra and (C) transmission electron micrographs (TEM) of the RGD-SERRS nanoparticles (red) and the non-targeted control RAD-SERRS nanoparticles (green). Scale bar = 100 nm. (D) Integrin-targeted SERRS nanoparticles enable the detection of bulk and infiltrative glioblastomas. GBM-bearing animals were co-injected with equimolar amounts of RGD-SERRS (targeted; red) and RAD-SERRS (non-targeted; green) nanoparticles. Multiplexed Raman imaging was performed on a coronal brain section (thickness 1 mm) in a paraffin block generated from a representative GBM-bearing mouse. Both RGD-SERRS (red) and RAD-SERRS (green) nanoparticles were detected in the bulk tumor located in the right hemisphere (OLIG2-positive). RGD-SERRS nanoparticles outlined the true tumor extent markedly better than the RAD-SERRS nanoparticles. (E) Sequential sections cut from the Raman-imaged paraffin block processed with H&E staining and immunohistochemistry of integrin β3 (α-ITGB3, target of the RGD-SERRS nanoparticles), OLIG2 (α-OLIG2), and polyethylene glycol linker (α-PEG, nanoparticle presence), respectively. (F) Pearson correlation analysis of the RGD-SERRS positive areas and OLIG-2 staining (n=7) was performed and a strong correlation (R=0.84) was found. Reproduced with permission from [151].
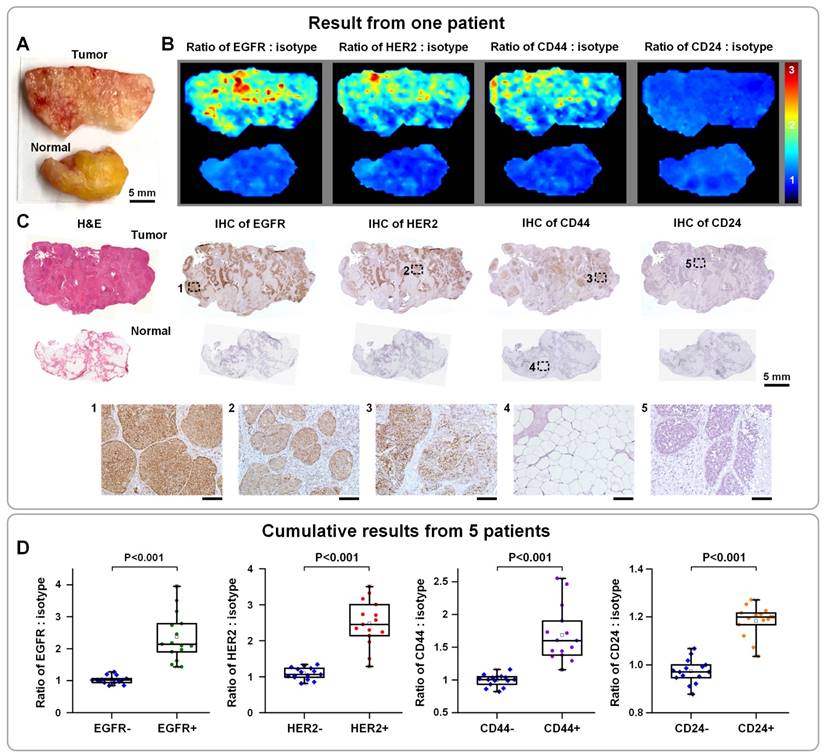
Multiplexed molecular imaging of freshly excised breast tissues with SERS NPs for guiding breast-conserving surgery. Each tissue specimen was stained with an equimolar mixture of HER2-NPs, EGFR-NPs, CD44-NPs, CD24-NPs and isotype-NPs (5 min), followed by a quick rinse in PBS (10 s) and raster-scanned imaging (< 3 min) to simultaneously quantify the expression of four breast cancer biomarkers: EGFR, HER2, CD44 and CD24. (A) Photograph of a human breast tumor and a normal tissue specimen from one patient. (B) Ratiometric images of EGFR-NPs vs. isotype-NPs, HER2-NPs vs. isotype-NPs, CD44-NPs vs. isotype-NPs and CD24-NPs. (C) Validation data: H&E and IHC for EGFR, HER2, CD44 and CD24. Unlabeled scale bars represent 200 μm. (D) Cumulative results from multiple regions of interest from a total of 5 patient specimens: measured NP ratios on IHC-validated biomarker-negative and biomarker-positive tissue regions. Each data point in the plots is the average ratio from one region of interest. Reproduced with permission from [62].
Molecular pathology
For over four decades, immunohistochemistry (IHC) analysis of FFPE tissue sections has been a clinical mainstay for the diagnosis, grading, and subtyping of tumors. However, IHC is semi-quantitative, subjective [157, 158], and, in the case of standard chromogenic staining methods, is unable to achieve multiplexed molecular imaging. Recently, many studies have shown that linear labeling and quantitative analysis of biomarkers in tissue sections can be achieved using bioconjugated nanoparticles such as QDs, with added benefits of high signal stability (no photobleaching) and a high degree of multiplexing [59, 60, 72, 159-164].
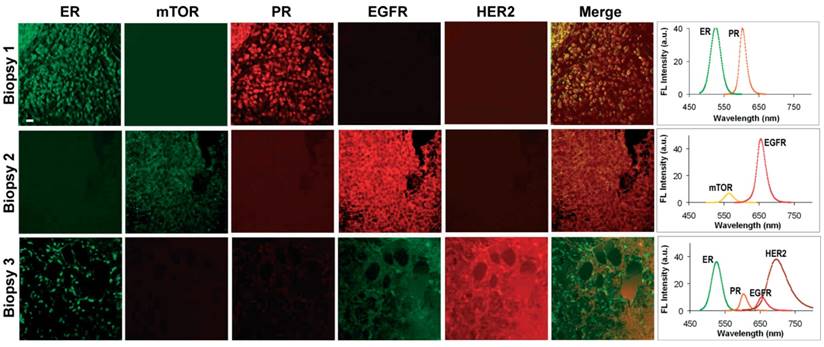
Simultaneous quantification of 5 biomarkers using QD-based IHC has been demonstrated on human prostate [159], breast [60] (Figure 10) and lymphoid tissue sections [72] as well as on mouse spleen tissue sections [164]. For image acquisition, the first study utilized a multispectral imaging system (Nuance, CRI) to scan a 580‒700 nm wavelength range using a LCTF (20-nm bandwidth, 13 channels at 10-nm increments). Detection was achieved with a cooled, scientific-grade monochrome CCD camera (overall acquisition time ~10 s per multispectral image)[159]. The second and third studies utilized a Zeiss LSM 510 META confocal microscope with a META detector [72].
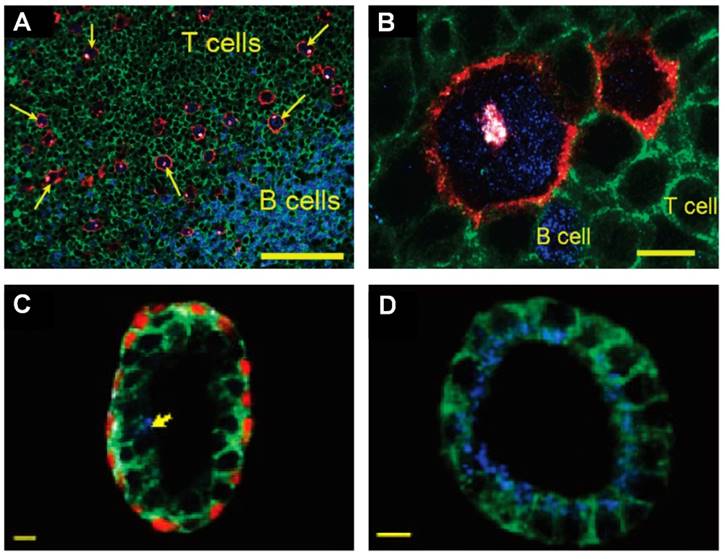
Multiplexed IHC using quantum dots could also facilitate the detection and characterization of tumor cells in complex tissue microenvironments [59, 165]. For example, multiplexed imaging of 4 biomarkers has been shown to enable the identification of rare (~1%) Hodgkins's and Reed-Sternberg (HRS) cells from infiltrating immune cells such as T and B lymphocytes (Figure 11A and 11B) [59]. This study also demonstrated a distinct QD staining pattern of HRS cells that can be used to differentiate Hodgkin's lymphoma from benign lymphoid hyperplasia. In another study, multiplexed imaging of 4 biomarkers using QDs was shown to allow detection of structurally distinct prostate glands and single cancer cells within the complex microenvironments of radical prostatectomy and needle biopsy specimens [165]. This study suggested that multiplexed IHC provides correlated molecular and morphological information that is not available from traditional IHC and H&E. The study also illustrated the progressive morphological and molecular changes of a benign prostate gland (double layer of basal and luminal cells with a single malignant cell, Figure 11C) to a completely malignant gland (a single layer of malignant cells, Figure 11D), indicating the convenience of multiplexed IHC for the study of tumor progression [165]. Similar to the system in [159], in these two studies, a multispectral imaging system (Nuance, CRI) was mounted on an inverted fluorescence microscope (Olympus IX71) for wavelength-resolved imaging. Improved understanding of tumor progression mechanisms and more rigorous classification can lead to more effective stage-specific and personalized treatments.
Summary and outlook
Molecular heterogeneity amongst tumors presents a challenge for early detection and treatment. Certain types of nanoparticles and imaging systems have been developed to enable multiplexed molecular imaging, which allows for effective imaging of cancers in spite of molecular heterogeneity. In this review, we briefly introduced the optical properties of major classes of nanoparticles that can be highly multiplexed such as QDs and SERS NPs, and summarized current imaging approaches and systems for the multiplexed imaging of these nanoparticles. In addition, by listing representative preclinical and clinical studies, we showcased some potential clinical applications of these multiplexed molecular imaging techniques such as for early detection of cancer, surgical guidance, and molecular pathology.
Although multiplexed molecular imaging techniques have grown significantly in popularity over the past few decades, there are clear opportunities to further improve their speed, accuracy and robustness, an essential step for their translation into clinical practice.
Multiplexed molecular profiling of FFPE tissue sections using QDs. Different patterns of nuclear, cytoplasmic and cell membrane fluorescent signals were detected by microscopy (left panels, pseudo-color) and expression of these biomarkers was quantified by wavelength-resolved spectroscopy (right panels). The emission spectra were resolved into individual channels and compensated for the differential brightness between different colors. Scale bar represents 20 µm. Reproduced with permission from [60].
QD-based IHC for molecular pathology. (A, B) Multispectral imaging of 4 QDs for detection of rare Hodgkin's and Reed-Sternberg (HRS) tumor cells in Hodgkin's lymphoma. The images show HRS malignant cells and infiltrating immune cells on lymph node tissue specimens. The HRS cells (arrows) exhibited a characteristic staining pattern: membrane staining (CD30 positive, red), Golgi staining (CD15 positive, white), and nuclear staining (Pax5 positive, blue). Staining patterns were clearly distinct from infiltrating B cells (blue nuclear staining) and T cells (green membrane staining). The scale bar in (A) represents 100 μm; the scale bar in (B) represents 10 μm. Reproduced with permission from [59]. Copyright (2010) American Chemical Society. (C, D) QD multiplexed staining of four protein biomarkers in human prostatectomy specimens highlighting cellular and glandular heterogeneity and tumor progression. The four biomarkers are E-cadherin (green), cytokeratin HMW (white), p63 (red) and AMACR (blue). (C) Largely benign prostate gland with a single malignant cell (arrow) in the luminal layer, as determined by positive AMACR staining. (D) Completely malignant gland, as determined by intense AMACR staining and absent basal cell cytokeratin and p63 signals. The scale bar in (C, D) represents 10 μm. Reproduced with permission from [165]. Copyright (2010) American Chemical Society.
Brighter nanoparticles are always desired to improve imaging speed, spatial resolution, and contrast (ratio of signal to tissue background). For example, some studies have reported polymer dots with a 10-fold enhanced brightness over commercial fluorescent dyes and QDs [15, 25], which can improve the imaging speed and image contrast, especially in the visible range where tissue autofluorescence signals are high. The relative weakness of SERS NP signals limits spectral acquisition rates and scanning speeds, which in turn limits the ability to scan large areas with high spatial resolution (i.e. imaging with large pixel counts). However, newly developed SERRS NPs have exhibited enhanced brightness over non-resonant SERS NPs, enabling the detection of small cancer-cell clusters with microscopic resolution [48, 166, 167].
Unlike label-free methods, molecular imaging based on exogenous agents involves complex interactions between contrast agents and tissue, both for delivery, retention, and wash out [168, 169]. In particular, systemically administered nanoparticles have been plagued by low delivery efficiency (~0.7% delivered to solid tumors) [168]. As an alternative approach, topically administered fluorescent dyes [169], QDs [170] and SERS NPs [75] have been used for molecular endoscopy and for the imaging of exposed surgical margins of freshly resected tissues. Topical application has been shown to be capable of achieving rapid and efficient biomarker labeling. An additional advantage is reduced toxicity and regulatory issues. For both systemic and topical administration, nonspecific accumulation and off-target binding of imaging probes in tissue specimens often confound the interpretation of imaging results. One way to normalize for those nonspecific effects is to utilize a ratiometric (or paired-agent) strategy in which a targeted probe and a control probe are simultaneously administered/applied such that the control probe serves as a quantitative control (and calibration factor) for all nonspecific effects. Compared to the conventional approach where only targeted probes are used, the ratiometric approach has been shown to improve biomarker quantification using imaging probes of various types such as fluorescent dyes [171-174] and SERS NPs [48, 62, 63, 65, 113, 154, 155](Figure 9). Note that SERS NPs are superior to fluorescent dyes for ratiometric imaging because different SERS NP flavors are identical in terms of geometry and surface properties (same coating materials), which ensures that nonspecific effects are identical for all NP flavors. Furthermore, all SERS NPs can be excited at a single wavelength, and imaged in the same wavelength range, which obviates wavelength-dependent effects. Finally, numerical modeling of probe delivery and retention kinetics has been demonstrated as a potential approach to accurately quantify biomarker expression levels based on the imaging of targeted fluorescent dyes and SERS NPs [156, 174].
With sustained efforts in nanoparticle chemistry, optics, biology, and other fields, we believe that multiplexed molecular imaging techniques will provide benefits for a variety of clinical applications in the future, such as for early cancer diagnosis, surgical guidance, and molecular pathology.
Acknowledgements
The authors acknowledge support from the NIH / NIBIB R21 EB015016 (J.T.C.L.), the NIH / NCI R21 CA215561 (J.T.C.L.), the Department of Mechanical Engineering at the University of Washington, and the department of education GAANN fellowship program (S.K.). The Northwest BioTrust (NWBT) is supported, in part, by the NIH (P30-CA015704).
Competing Interests
Jonathan T.C. Liu is an inventor for a pending patent: “Raman imaging devices and methods of molecular imaging.” The other authors have declared that no conflict of interest exists.
References
1. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M. et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-E86
2. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90
3. Rapiti E, Verkooijen HM, Vlastos G, Fioretta G, Neyroud-Caspar I, Sappino AP. et al. Complete Excision of Primary Breast Tumor Improves Survival of Patients With Metastatic Breast Cancer at Diagnosis. J Clin Oncol. 2006;24:2743-9
4. Weissleder R, Ross BD, Rehemtulla A, Gambhir SS. Molecular imaging: principles and practice. People's Medical Publishing House, USA. 2010
5. Hussain T, Nguyen QT. Molecular imaging for cancer diagnosis and surgery. Adv Drug Del Rev. 2014;66:90-100
6. Mankoff DA, Pryma DA, Clark AS. Molecular imaging biomarkers for oncology clinical trials. J Nucl Med. 2014;55:525-8
7. Pysz MA, Gambhir SS, Willmann JK. Molecular imaging: current status and emerging strategies. Clin Radiol. 2010;65:500-16
8. Fass L. Imaging and cancer: a review. Mol Oncol. 2008;2:115-52
9. Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochim Biophys Acta. 2010;1805:105-17
10. Almendro V, Marusyk A, Polyak K. Cellular heterogeneity and molecular evolution in cancer. Annu Rev Pathol. 2013;8:277-302
11. Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328-37
12. Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E. et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. N Engl J Med. 2012;366:883-92
13. Kobayashi H, Ogawa M, Alford R, Choyke PL, Urano Y. New Strategies for Fluorescent Probe Design in Medical Diagnostic Imaging. Chem Rev. 2010;110:2620-40
14. Resch-Genger U, Grabolle M, Cavaliere-Jaricot S, Nitschke R, Nann T. Quantum dots versus organic dyes as fluorescent labels. Nat Meth. 2008;5:763-75
15. Wu C, Chiu DT. Highly Fluorescent Semiconducting Polymer Dots for Biology and Medicine. Angewandte Chemie International Edition. 2013;52:3086-109
16. Ntziachristos V, Bremer C, Weissleder R. Fluorescence imaging with near-infrared light: new technological advances that enable in vivo molecular imaging. Eur Radiol. 2003;13:195-208
17. Zrazhevskiy P, Sena M, Gao X. Designing multifunctional quantum dots for bioimaging, detection, and drug delivery. Chem Soc Rev. 2010;39:4326-54
18. Montalti M, Prodi L, Rampazzo E, Zaccheroni N. Dye-doped silica nanoparticles as luminescent organized systems for nanomedicine. Chem Soc Rev. 2014;43:4243-68
19. Wang L, Wang K, Santra S, Zhao X, Hilliard LR, Smith JE. et al. Watching Silica Nanoparticles Glow in the Biological World. Anal Chem. 2006;78:646-54
20. Zhao X, Bagwe RP, Tan W. Development of Organic-Dye-Doped Silica Nanoparticles in a Reverse Microemulsion. Advanced Materials. 2004;16:173-6
21. Bae SW, Tan W, Hong J-I. Fluorescent dye-doped silica nanoparticles: new tools for bioapplications. Chemical Communications. 2012;48:2270-82
22. Wang L, Tan W. Multicolor FRET Silica Nanoparticles by Single Wavelength Excitation. Nano Lett. 2006;6:84-8
23. Chen X, Estévez MC, Zhu Z, Huang Y-F, Chen Y, Wang L. et al. Using Aptamer-Conjugated Fluorescence Resonance Energy Transfer Nanoparticles for Multiplexed Cancer Cell Monitoring. Anal Chem. 2009;81:7009-14
24. Alivisatos AP. Semiconductor Clusters, Nanocrystals, and Quantum Dots. Science. 1996;271:933-7
25. Yu J, Rong Y, Kuo C-T, Zhou X-H, Chiu DT. Recent Advances in the Development of Highly Luminescent Semiconducting Polymer Dots and Nanoparticles for Biological Imaging and Medicine. Anal Chem. 2017;89:42-56
26. Wu IC, Yu J, Ye F, Rong Y, Gallina ME, Fujimoto BS. et al. Squaraine-based polymer dots with narrow, bright near-infrared fluorescence for biological applications. J Am Chem Soc. 2015;137:173-8
27. Rong Y, Wu C, Yu J, Zhang X, Ye F, Zeigler M. et al. Multicolor Fluorescent Semiconducting Polymer Dots with Narrow Emissions and High Brightness. ACS Nano. 2013;7:376-84
28. Foubert A, Beloglazova NV, Rajkovic A, Sas B, Madder A, Goryacheva IY. et al. Bioconjugation of quantum dots: Review & impact on future application. TrAC Trends in Analytical Chemistry. 2016;83(Part B):31-48
29. Esteve-Turrillas FA, Abad-Fuentes A. Applications of quantum dots as probes in immunosensing of small-sized analytes. Biosens Bioelectron. 2013;41:12-29
30. Chan WC, Nie S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science. 1998;281:2016-8
31. Bruchez M Jr, Moronne M, Gin P, Weiss S, Alivisatos AP. Semiconductor nanocrystals as fluorescent biological labels. Science. 1998;281:2013-6
32. Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ. et al. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307:538-44
33. Dabbousi BO, Rodriguez-Viejo J, Mikulec FV, Heine JR, Mattoussi H, Ober R. et al. (CdSe)ZnS Core-Shell Quantum Dots: Synthesis and Characterization of a Size Series of Highly Luminescent Nanocrystallites. The Journal of Physical Chemistry B. 1997;101:9463-75
34. Nie S, Emory SR. Probing Single Molecules and Single Nanoparticles by Surface-Enhanced Raman Scattering. Science. 1997;275:1102-6
35. Haynes CL, McFarland AD, Duyne RPV. Surface-Enhanced Raman Spectroscopy. Anal Chem. 2005;77:338A-46A
36. Sonntag MD, Klingsporn JM, Zrimsek AB, Sharma B, Ruvuna LK, Van Duyne RP. Molecular plasmonics for nanoscale spectroscopy. Chem Soc Rev. 2014;43:1230-47
37. Kneipp K, Moskovits M, Kneipp H. Surface-Enhanced Raman Scattering: Physics and Applications. New York: Springer Berlin Heidelberg. 2006
38. Fleischmann M, Hendra PJ, McQuillan AJ. Raman spectra of pyridine adsorbed at a silver electrode. Chem Phys Lett. 1974;26:163-6
39. Albrecht MG, Creighton JA. Anomalously intense Raman spectra of pyridine at a silver electrode. J Am Chem Soc. 1977;99:5215-7
40. Jeanmaire DL, Van Duyne RP. Surface raman spectroelectrochemistry. Journal of Electroanalytical Chemistry and Interfacial Electrochemistry. 1977;84:1-20
41. Kneipp K, Wang Y, Kneipp H, Perelman LT, Itzkan I, Dasari RR. et al. Single Molecule Detection Using Surface-Enhanced Raman Scattering (SERS). Phys Rev Lett. 1997;78:1667-70
42. Gruenke NL, Cardinal MF, McAnally MO, Frontiera RR, Schatz GC, Van Duyne RP. Ultrafast and nonlinear surface-enhanced Raman spectroscopy. Chem Soc Rev. 2016;45:2263-90
43. Hering K, Cialla D, Ackermann K, Dorfer T, Moller R, Schneidewind H. et al. SERS: a versatile tool in chemical and biochemical diagnostics. Anal Bioanal Chem. 2008;390:113-24
44. Stiles PL, Dieringer JA, Shah NC, Van Duyne RP. Surface-enhanced Raman spectroscopy. Annu Rev Anal Chem. 2008;1:601-26
45. Kneipp K, Kneipp H, Kneipp J. Surface-Enhanced Raman Scattering in Local Optical Fields of Silver and Gold NanoaggregatesFrom Single-Molecule Raman Spectroscopy to Ultrasensitive Probing in Live Cells. Acc Chem Res. 2006;39:443-50
46. Wang Y, Yan B, Chen L. SERS tags: novel optical nanoprobes for bioanalysis. Chem Rev. 2013;113:1391-428
47. Austin LA, Osseiran S, Evans CL. Raman technologies in cancer diagnostics. Analyst. 2016;141:476-503
48. Oseledchyk A, Andreou C, Wall MA, Kircher MF. Folate-Targeted Surface-Enhanced Resonance Raman Scattering Nanoprobe Ratiometry for Detection of Microscopic Ovarian Cancer. ACS Nano. 2017;11:1488-97
49. Harmsen S, Bedics MA, Wall MA, Huang R, Detty MR, Kircher MF. Rational design of a chalcogenopyrylium-based surface-enhanced resonance Raman scattering nanoprobe with attomolar sensitivity. Nature Communications. 2015;6:6570
50. Wang Y, Schlucker S. Rational design and synthesis of SERS labels. Analyst. 2013;138:2224-38
51. Qian X, Peng XH, Ansari DO, Yin-Goen Q, Chen GZ, Shin DM. et al. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nat Biotechnol. 2008;26:83-90
52. Zavaleta CL, Garai E, Liu JT, Sensarn S, Mandella MJ, Van de Sompel D. et al. A Raman-based endoscopic strategy for multiplexed molecular imaging. Proc Natl Acad Sci U S A. 2013;110:E2288-97
53. Zavaleta CL, Smith BR, Walton I, Doering W, Davis G, Shojaei B. et al. Multiplexed imaging of surface enhanced Raman scattering nanotags in living mice using noninvasive Raman spectroscopy. Proc Natl Acad Sci U S A. 2009;106:13511-6
54. Doering WE, Nie S. Spectroscopic Tags Using Dye-Embedded Nanoparticles and Surface-Enhanced Raman Scattering. Anal Chem. 2003;75:6171-6
55. Liu Z, Tabakman S, Sherlock S, Li X, Chen Z, Jiang K. et al. Multiplexed five-color molecular imaging of cancer cells and tumor tissues with carbon nanotube Raman tags in the near-infrared. Nano Research. 2010;3:222-33
56. Leblond F, Davis SC, Valdés PA, Pogue BW. Pre-clinical whole-body fluorescence imaging: Review of instruments, methods and applications. J Photochem Photobiol B: Biol. 2010;98:77-94
57. Kobayashi H, Hama Y, Koyama Y, Barrett T, Regino CAS, Urano Y. et al. Simultaneous Multicolor Imaging of Five Different Lymphatic Basins Using Quantum Dots. Nano Lett. 2007;7:1711-6
58. Zrazhevskiy P, Gao X. Quantum dot imaging platform for single-cell molecular profiling. Nature Communications. 2013;4:1619
59. Liu J, Lau SK, Varma VA, Kairdolf BA, Nie S. Multiplexed detection and characterization of rare tumor cells in Hodgkin's lymphoma with multicolor quantum dots. Anal Chem. 2010;82:6237-43
60. Yezhelyev MV, Al-Hajj A, Morris C, Marcus AI, Liu T, Lewis M. et al. In Situ Molecular Profiling of Breast Cancer Biomarkers with Multicolor Quantum Dots. Advanced Materials. 2007;19:3146-51
61. Muldoon TJ, Pierce MC, Nida DL, Williams MD, Gillenwater A, Richards-Kortum R. Subcellular-resolution molecular imaging within living tissue by fiber microendoscopy. Opt Express. 2007;15:16413-23
62. Wang YW, Doerksen JD, Kang S, Walsh D, Yang Q, Hong D. et al. Multiplexed Molecular Imaging of Fresh Tissue Surfaces Enabled by Convection-Enhanced Topical Staining with SERS-Coded Nanoparticles. Small (Weinheim an der Bergstrasse, Germany). 2016;12:5612-21
63. Wang Y, Reder NP, Kang S, Glaser AK, Yang Q, Wall MA. et al. Raman-encoded molecular imaging (REMI) with topically applied SERS nanoparticles for intraoperative guidance of lumpectomy. Cancer Res. 2017
64. McVeigh PZ, Mallia RJ, Veilleux I, Wilson BC. Widefield quantitative multiplex surface enhanced Raman scattering imaging in vivo. J Biomed Opt. 2013;18:046011 -
65. Mallia RJ, McVeigh PZ, Fisher CJ, Veilleux I, Wilson BC. Wide-field multiplexed imaging of EGFR-targeted cancers using topical application of NIR SERS nanoprobes. Nanomedicine (Lond). 2015;10:89-101
66. Zimmermann T, Marrison J, Hogg K, O'Toole P. Clearing up the signal: spectral imaging and linear unmixing in fluorescence microscopy. Methods Mol Biol. 2014;1075:129-48
67. Goldman ER, Clapp AR, Anderson GP, Uyeda HT, Mauro JM, Medintz IL. et al. Multiplexed Toxin Analysis Using Four Colors of Quantum Dot Fluororeagents. Anal Chem. 2004;76:684-8
68. Levenson RM, Mansfield JR. Multispectral imaging in biology and medicine: Slices of life. Cytometry Part A. 2006;69A:748-58
69. Valm AM, Oldenbourg R, Borisy GG. Multiplexed Spectral Imaging of 120 Different Fluorescent Labels. PLoS One. 2016;11:e0158495
70. Van de Sompel D, Garai E, Zavaleta C, Gambhir SS. A Hybrid Least Squares and Principal Component Analysis Algorithm for Raman Spectroscopy. PLoS One. 2012;7:e38850
71. Zimmermann T. Spectral Imaging and Linear Unmixing in Light Microscopy. In: (ed.) Rietdorf J. Microscopy Techniques: -/-. Berlin, Heidelberg: Springer Berlin Heidelberg. 2005:245-65
72. Fountaine TJ, Wincovitch SM, Geho DH, Garfield SH, Pittaluga S. Multispectral imaging of clinically relevant cellular targets in tonsil and lymphoid tissue using semiconductor quantum dots. Mod Pathol. 2006;19:1181-91
73. Garini Y, Young IT, McNamara G. Spectral imaging: Principles and applications. Cytometry Part A. 2006;69A:735-47
74. Petryayeva E, Algar WR, Medintz IL. Quantum Dots in Bioanalysis: A Review of Applications across Various Platforms for Fluorescence Spectroscopy and Imaging. Applied Spectroscopy. 2013;67:215-52
75. Wang Y, Kang S, Doerksen J, Glaser A, Liu J. Surgical Guidance via Multiplexed Molecular Imaging of Fresh Tissues Labeled with SERS-Coded Nanoparticles. IEEE J Sel Top Quantum Electron. 2016;22:154-64
76. Mansfield JR, Hoyt C, Levenson RM. Visualization of microscopy-based spectral imaging data from multi-label tissue sections. Curr Protoc Mol Biol. 2008 Chapter 14: Unit 14.9
77. Neher R, Neher E. Optimizing imaging parameters for the separation of multiple labels in a fluorescence image. J Microsc. 2004;213:46-62
78. Kong K, Rowlands CJ, Varma S, Perkins W, Leach IH, Koloydenko AA. et al. Diagnosis of tumors during tissue-conserving surgery with integrated autofluorescence and Raman scattering microscopy. Proc Natl Acad Sci U S A. 2013;110:15189-94
79. Han X, Lui H, McLean DI, Zeng H. Near-infrared autofluorescence imaging of cutaneous melanins and human skin in vivo. J Biomed Opt. 2009;14:024017
80. Lutz BR, Dentinger CE, Nguyen LN, Sun L, Zhang J, Allen AN. et al. Spectral Analysis of Multiplex Raman Probe Signatures. ACS Nano. 2008;2:2306-14
81. Zhao J, Lui H, McLean DI, Zeng H. Automated autofluorescence background subtraction algorithm for biomedical Raman spectroscopy. Appl Spectrosc. 2007;61:1225-32
82. Lieber CA, Mahadevan-Jansen A. Automated Method for Subtraction of Fluorescence from Biological Raman Spectra. Applied Spectroscopy. 2003;57:1363-7
83. Zhang ZM, Chen S, Liang YZ. Baseline correction using adaptive iteratively reweighted penalized least squares. Analyst. 2010;135:1138-46
84. Kode K, Shachaf C, Elchuri S, Nolan G, Paik DS. Raman labeled nanoparticles: characterization of variability and improved method for unmixing. J Raman Spectrosc. 2012;43:895-905
85. Garai E, Sensarn S, Zavaleta CL, Van de Sompel D, Loewke NO, Mandella MJ. et al. High-sensitivity, real-time, ratiometric imaging of surface-enhanced Raman scattering nanoparticles with a clinically translatable Raman endoscope device. J Biomed Opt. 2013;18:096008
86. Paddock SW, Eliceiri KW. Laser Scanning Confocal Microscopy: History, Applications, and Related Optical Sectioning Techniques. In: (ed.) Paddock SW. Confocal Microscopy: Methods and Protocols. New York, NY: Springer New York. 2014:9-47
87. Lee CM, Engelbrecht CJ, Soper TD, Helmchen F, Seibel EJ. Scanning fiber endoscopy with highly flexible, 1 mm catheterscopes for wide-field, full-color imaging. Journal of biophotonics. 2010;3:385-407
88. Rogalla S, Contag CH. Early Cancer Detection at the Epithelial Surface. The Cancer Journal. 2015;21:179-87
89. Lawrence WG, Varadi G, Entine G, Podniesinski E, Wallace PK. A comparison of avalanche photodiode and photomultiplier tube detectors for flow cytometry. 2008: 68590M-M-11.
90. Maiti KK, Dinish US, Samanta A, Vendrell M, Soh K-S, Park S-J. et al. Multiplex targeted in vivo cancer detection using sensitive near-infrared SERS nanotags. Nano Today. 2012;7:85-93
91. Glaser AK, Wang Y, Liu JT. Assessing the imaging performance of light sheet microscopies in highly scattering tissues. Biomed Opt Express. 2016;7:454-66
92. Kang S, Wang Y, Reder NP, Liu JTC. Multiplexed Molecular Imaging of Biomarker-Targeted SERS Nanoparticles on Fresh Tissue Specimens with Channel-Compressed Spectrometry. PLoS One. 2016;11:e0163473
93. Gustafsson MGL. Nonlinear structured-illumination microscopy: Wide-field fluorescence imaging with theoretically unlimited resolution. Proc Natl Acad Sci U S A. 2005;102:13081-6
94. Fereidouni F, Datta-Mitra A, Demos S, Levenson R. Microscopy with UV Surface Excitation (MUSE) for slide-free histology and pathology imaging. 2015. p. 93180F-F-6.
95. Wang M, Kimbrell HZ, Sholl AB, Tulman DB, Elfer KN, Schlichenmeyer TC. et al. High-Resolution Rapid Diagnostic Imaging of Whole Prostate Biopsies Using Video-Rate Fluorescence Structured Illumination Microscopy. Cancer Res. 2015;75:4032-41
96. Schlichenmeyer TC, Wang M, Elfer KN, Brown JQ. Video-rate structured illumination microscopy for high-throughput imaging of large tissue areas. Biomed Opt Express. 2014;5:366-77
97. Neil MAA, Juškaitis R, Wilson T. Method of obtaining optical sectioning by using structured light in a conventional microscope. Optics letters. 1997;22:1905-7
98. Karadaglić D, Wilson T. Image formation in structured illumination wide-field fluorescence microscopy. Micron. 2008;39:808-18
99. Dwight JG, Tkaczyk TS. Lenslet array tunable snapshot imaging spectrometer (LATIS) for hyperspectral fluorescence microscopy. Biomed Opt Express. 2017;8:1950-64
100. Lu G, Fei B. Medical hyperspectral imaging: a review. J Biomed Opt. 2014;19:010901 -
101. Gao L, Wang LV. A review of snapshot multidimensional optical imaging: Measuring photon tags in parallel. Phys Rep. 2016;616:1-37
102. Sun D-W. Hyperspectral imaging for food quality analysis and control. Elsevier. 2010
103. Zhou L, El-Deiry WS. Multispectral fluorescence imaging. J Nucl Med. 2009;50:1563-6
104. Gao L, Kester RT, Hagen N, Tkaczyk TS. Snapshot Image Mapping Spectrometer (IMS) with high sampling density for hyperspectral microscopy. Opt Express. 2010;18:14330-44
105. Kester RT, Bedard N, Gao L, Tkaczyk TS. Real-time snapshot hyperspectral imaging endoscope. J Biomed Opt. 2011:16
106. Hagen N, Kester RT, Gao L, Tkaczyk TS. Snapshot advantage: a review of the light collection improvement for parallel high-dimensional measurement systems. OPTICE. 2012;51:111702-1 -7
107. Gao L, Smith RT. Optical hyperspectral imaging in microscopy and spectroscopy - a review of data acquisition. Journal of biophotonics. 2015;8:441-56
108. Bedard N, Schwarz RA, Hu A, Bhattar V, Howe J, Williams MD. et al. Multimodal snapshot spectral imaging for oral cancer diagnostics: a pilot study. Biomed Opt Express. 2013;4:938-49
109. Zhu H, Isikman SO, Mudanyali O, Greenbaum A, Ozcan A. Optical imaging techniques for point-of-care diagnostics. Lab on a Chip. 2013;13:51-67
110. Oh G, Chung E, Yun SH. Optical fibers for high-resolution in vivo microendoscopic fluorescence imaging. Optical Fiber Technology. 2013;19:760-71
111. Flusberg BA, Cocker ED, Piyawattanametha W, Jung JC, Cheung ELM, Schnitzer MJ. Fiber-optic fluorescence imaging. Nat Meth. 2005;2:941-50
112. Joshi BP, Miller SJ, Lee CM, Seibel EJ, Wang TD. Multispectral endoscopic imaging of colorectal dysplasia in vivo. Gastroenterology. 2012;143:1435-7
113. Wang YW, Kang S, Khan A, Bao PQ, Liu JTC. In vivo multiplexed molecular imaging of esophageal cancer via spectral endoscopy of topically applied SERS nanoparticles. Biomed Opt Express. 2015;6:3714-23
114. Yin C, Glaser AK, Leigh SY, Chen Y, Wei L, Pillai PCS. et al. Miniature in vivo MEMS-based line-scanned dual-axis confocal microscope for point-of-care pathology. Biomed Opt Express. 2016;7:251-63
115. Ra H, Piyawattanametha W, Gonzalez-Gonzalez E, Mandella MJ, Kino GS, Solgaard O. et al. In Vivo Imaging of Human and Mouse Skin with a Handheld Dual-Axis Confocal Fluorescence Microscope. The Journal of investigative dermatology. 2011;131:10.1038 /jid.2010.401
116. Matsuoka H, Kosai Y, Saito M, Takeyama N, Suto H. Single-cell viability assessment with a novel spectro-imaging system. J Biotechnol. 2002;94:299-308
117. Fletcher-Holmes DW, Harvey AR. Real-time imaging with a hyperspectral fovea. Journal of Optics A: Pure and Applied Optics. 2005;7:S298
118. Gat N, Scriven G, Garman J, De Li M, Zhang J. Development of four-dimensional imaging spectrometers (4D-IS). SPIE Optics+ Photonics: International Society for Optics and Photonics. 2006 p. 63020M-M-11
119. Kriesel J, Scriven G, Gat N, Nagaraj S, Willson P, Swaminathan V. Snapshot hyperspectral fovea vision system (HyperVideo). SPIE Defense, Security, and Sensing: International Society for Optics and Photonics. 2012 p. 83900T-T-6
120. Shadfan A, Darwiche H, Blanco J, Gillenwater A, Richards-Kortum R, Tkaczyk TS. Development of a multimodal foveated endomicroscope for the detection of oral cancer. Biomed Opt Express. 2017;8:1525-35
121. Li Q, He X, Wang Y, Liu H, Xu D, Guo F. Review of spectral imaging technology in biomedical engineering: achievements and challenges. J Biomed Opt. 2013;18:100901 -
122. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7-30
123. Barenholz Y. Doxil® — The first FDA-approved nano-drug: Lessons learned. J Control Release. 2012;160:117-34
124. Biffi S, Voltan R, Rampazzo E, Prodi L, Zauli G, Secchiero P. Applications of nanoparticles in cancer medicine and beyond: optical and multimodal in vivo imaging, tissue targeting and drug delivery. Expert Opinion on Drug Delivery. 2015;12:1837-49
125. Miller SJ, Lee CM, Joshi BP, Gaustad A, Seibel EJ, Wang TD. Targeted detection of murine colonic dysplasia in vivo with flexible multispectral scanning fiber endoscopy. J Biomed Opt. 2012;17:021103
126. Wang YW, Khan A, Leigh SY, Wang D, Chen Y, Meza D. et al. Comprehensive spectral endoscopy of topically applied SERS nanoparticles in the rat esophagus. Biomed Opt Express. 2014;5:2883-95
127. Garai E, Sensarn S, Zavaleta CL, Loewke NO, Rogalla S, Mandella MJ. et al. A real-time clinical endoscopic system for intraluminal, multiplexed imaging of surface-enhanced Raman scattering nanoparticles. PLoS One. 2015;10:e0123185
128. Short MA, Lam S, McWilliams A, Zhao J, Lui H, Zeng H. Development and preliminary results of an endoscopic Raman probe for potential in vivo diagnosis of lung cancers. Optics letters. 2008;33:711-3
129. Thakor AS, Luong R, Paulmurugan R, Lin FI, Kempen P, Zavaleta C. et al. The fate and toxicity of Raman-active silica-gold nanoparticles in mice. Sci Transl Med. 2011;3:79ra33
130. Zavaleta CL, Hartman KB, Miao Z, James ML, Kempen P, Thakor AS. et al. Preclinical evaluation of Raman nanoparticle biodistribution for their potential use in clinical endoscopy imaging. Small (Weinheim an der Bergstrasse, Germany). 2011;7:2232-40
131. Campbell JL, SoRelle ED, Ilovich O, Liba O, James ML, Qiu Z. et al. Multimodal assessment of SERS nanoparticle biodistribution post ingestion reveals new potential for clinical translation of Raman imaging. Biomaterials. 2017
132. Jacobs L. Positive margins: the challenge continues for breast surgeons. Ann Surg Oncol. 2008;15:1271-2
133. Yossepowitch O, Bjartell A, Eastham JA, Graefen M, Guillonneau BD, Karakiewicz PI. et al. Positive surgical margins in radical prostatectomy: outlining the problem and its long-term consequences. Eur Urol. 2009;55:87-99
134. Ang P, Tan AW, Goh CL. Comparison of completely versus incompletely excised cutaneous squamous cell carcinomas. Ann Acad Med Singapore. 2004;33:68-70
135. Talbot S, Hitchcock B. Incomplete primary excision of cutaneous basal and squamous cell carcinomas in the Bay of Plenty. N Z Med J. 2004;117:U848
136. Haque R, Contreras R, McNicoll MP, Eckberg EC, Petitti DB. Surgical margins and survival after head and neck cancer surgery. BMC Ear Nose Throat Disord. 2006;6:2
137. Pogue BW, Paulsen KD, Samkoe KS, Elliott JT, Hasan T, Strong TV. et al. Vision 20/20: Molecular-guided surgical oncology based upon tumor metabolism or immunologic phenotype: Technological pathways for point of care imaging and intervention. Med Phys. 2016;43:3143-56
138. Bu L, Shen B, Cheng Z. Fluorescent imaging of cancerous tissues for targeted surgery. Adv Drug Del Rev. 2014;76:21-38
139. Adamson C, Kanu OO, Mehta AI, Di C, Lin N, Mattox AK. et al. Glioblastoma multiforme: a review of where we have been and where we are going. Expert opinion on investigational drugs. 2009;18:1061-83
140. Bucci MK, Maity A, Janss AJ, Belasco JB, Fisher MJ, Tochner ZA. et al. Near complete surgical resection predicts a favorable outcome in pediatric patients with nonbrainstem, malignant gliomas: results from a single center in the magnetic resonance imaging era. Cancer. 2004;101:817-24
141. Laura B. Ngwenya, E. Antonio Chiocca. Editorial: Extent of resection. J Neurosurg. 2011;115:1-2
142. Pogue BW, Gibbs-Strauss S, Valdés PA, Samkoe K, Roberts DW, Paulsen KD. Review of Neurosurgical Fluorescence Imaging Methodologies. IEEE journal of selected topics in quantum electronics: a publication of the IEEE Lasers and Electro-optics Society. 2010;16:493-505
143. Li Y, Rey-Dios R, Roberts DW, Valdés PA, Cohen-Gadol AA. Intraoperative Fluorescence-Guided Resection of High-Grade Gliomas: A Comparison of the Present Techniques and Evolution of Future Strategies. World Neurosurg. 2014;82:175-85
144. Arndt-Jovin DJ, Kantelhardt SR, Caarls W, Vries AHBd, Giese A, Jovin TM. et al. Tumor-Targeted Quantum Dots Can Help Surgeons Find Tumor Boundaries. IEEE Trans NanoBiosci. 2009;8:65-71
145. Cai W, Shin D-W, Chen K, Gheysens O, Cao Q, Wang SX. et al. Peptide-Labeled Near-Infrared Quantum Dots for Imaging Tumor Vasculature in Living Subjects. Nano Lett. 2006;6:669-76
146. Jackson H, Muhammad O, Daneshvar H, Nelms J, Popescu A, Vogelbaum MA. et al. Quantum dots are phagocytized by macrophages and colocalize with experimental gliomas. Neurosurgery. 2007;60:524-9 discussion 9-30
147. Jung J, Solanki A, Memoli KA, Kamei K, Kim H, Drahl MA. et al. Selective inhibition of human brain tumor cells through multifunctional quantum-dot-based siRNA delivery. Angew Chem Int Ed Engl. 2010;49:103-7
148. Wang J, Yong WH, Sun Y, Vernier PT, Koeffler HP, Gundersen MA. et al. Receptor-targeted quantum dots: fluorescent probes for brain tumor diagnosis. J Biomed Opt. 2007;12:044021-7
149. Kircher MF, de la Zerda A, Jokerst JV, Zavaleta CL, Kempen PJ, Mittra E. et al. A Brain Tumor Molecular Imaging Strategy Using A New Triple-Modality MRI-Photoacoustic-Raman Nanoparticle. Nat Med. 2012;18:829-34
150. Karabeber H, Huang R, Iacono P, Samii JM, Pitter K, Holland EC. et al. Guiding Brain Tumor Resection Using Surface-Enhanced Raman Scattering Nanoparticles and a Hand-Held Raman Scanner. ACS Nano. 2014;8:9755-66
151. Huang R, Harmsen S, Samii JM, Karabeber H, Pitter KL, Holland EC. et al. High Precision Imaging of Microscopic Spread of Glioblastoma with a Targeted Ultrasensitive SERRS Molecular Imaging Probe. Theranostics. 2016;6:1075-84
152. DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64:52-62
153. Gray RJ, Pockaj BA, Garvey E, Blair S. Intraoperative Margin Management in Breast-Conserving Surgery: A Systematic Review of the Literature. Ann Surg Oncol. 2017:1-10
154. Wang Y, Kang S, Khan A, Ruttner G, Leigh SY, Murray M. et al. Quantitative molecular phenotyping with topically applied SERS nanoparticles for intraoperative guidance of breast cancer lumpectomy. Sci Rep. 2016;6:21242
155. Wang YW, Khan A, Som M, Wang D, Chen Y, Leigh SY. et al. Rapid ratiometric biomarker detection with topically applied SERS nanoparticles. Technology. 2014;2:118-32
156. Sinha L, Wang Y, Yang C, Khan A, Brankov JG, Liu JTC. et al. Quantification of the binding potential of cell-surface receptors in fresh excised specimens via dual-probe modeling of SERS nanoparticles. Sci Rep. 2015;5:8582
157. Taylor CR, Levenson RM. Quantification of immunohistochemistry—issues concerning methods, utility and semiquantitative assessment II. Histopathology. 2006;49:411-24
158. Thomson TA, Hayes MM, Spinelli JJ, Hilland E, Sawrenko C, Phillips D. et al. HER-2/neu in breast cancer: interobserver variability and performance of immunohistochemistry with 4 antibodies compared with fluorescent in situ hybridization. Mod Pathol. 2001;14:1079-86
159. Xing Y, Chaudry Q, Shen C, Kong KY, Zhau HE, Chung LW. et al. Bioconjugated quantum dots for multiplexed and quantitative immunohistochemistry. Nat Protocols. 2007;2:1152-65
160. Chen C, Peng J, Xia HS, Yang GF, Wu QS, Chen LD. et al. Quantum dots-based immunofluorescence technology for the quantitative determination of HER2 expression in breast cancer. Biomaterials. 2009;30:2912-8
161. Chen H, Xue J, Zhang Y, Zhu X, Gao J, Yu B. Comparison of quantum dots immunofluorescence histochemistry and conventional immunohistochemistry for the detection of caveolin-1 and PCNA in the lung cancer tissue microarray. Journal of molecular histology. 2009;40:261-8
162. Chen C, Peng J, Xia H, Wu Q, Zeng L, Xu H. et al. Quantum-dot-based immunofluorescent imaging of HER2 and ER provides new insights into breast cancer heterogeneity. Nanotechnology. 2010;21:095101
163. Chen SQ, Zhang HM, Li JB, Jiang HY, Fan L, Kong LZ. et al. Analyzing simultaneous positive expression of EZH2 and P53 protein to improve predictive value in cervical squamous cell carcinoma. Int J Gynecol Cancer. 2014;24:1653-8
164. Jennings TL, Becker-Catania SG, Triulzi RC, Tao G, Scott B, Sapsford KE. et al. Reactive Semiconductor Nanocrystals for Chemoselective Biolabeling and Multiplexed Analysis. ACS Nano. 2011;5:5579-93
165. Liu J, Lau SK, Varma VA, Moffitt RA, Caldwell M, Liu T. et al. Molecular Mapping of Tumor Heterogeneity on Clinical Tissue Specimens with Multiplexed Quantum Dots. ACS nano. 2010;4:2755-65
166. Harmsen S, Huang R, Wall MA, Karabeber H, Samii JM, Spaliviero M. et al. Surface-Enhanced Resonance Raman Scattering Nanostars for High Precision Cancer Imaging. Sci Transl Med. 2015;7:271ra7-ra7
167. Spaliviero M, Harmsen S, Huang R, Wall MA, Andreou C, Eastham JA. et al. Detection of Lymph Node Metastases with SERRS Nanoparticles. Mol Imaging Biol. 2016;18:677-85
168. Wilhelm S, Tavares AJ, Dai Q, Ohta S, Audet J, Dvorak HF. et al. Analysis of nanoparticle delivery to tumours. Nature Reviews Materials. 2016;1:16014
169. Hellebust A, Richards-Kortum R. Advances in molecular imaging: targeted optical contrast agents for cancer diagnostics. Nanomedicine (London, England). 2012;7:429-45
170. Park Y, Ryu Y-M, Jung Y, Wang T, Baek Y, Yoon Y. et al. Spraying Quantum Dot Conjugates in the Colon of Live Animals Enabled Rapid and Multiplex Cancer Diagnosis Using Endoscopy. ACS Nano. 2014;8:8896-910
171. Tichauer KM, Samkoe KS, Gunn JR, Kanick SC, Hoopes PJ, Barth RJ. et al. Microscopic lymph node tumor burden quantified by macroscopic dual-tracer molecular imaging. Nat Med. 2014;20:1348-53
172. Tichauer KM, Samkoe KS, Sexton KJ, Hextrum SK, Yang HH, Klubben WS. et al. In vivo quantification of tumor receptor binding potential with dual-reporter molecular imaging. Mol Imaging Biol. 2012;14:584-92
173. Liu JTC, Helms MW, Mandella MJ, Crawford JM, Kino GS, Contag CH. Quantifying Cell-Surface Biomarker Expression in Thick Tissues with Ratiometric Three-Dimensional Microscopy. Biophys J. 2009;96:2405-14
174. Tichauer KM, Wang Y, Pogue BW, Liu JTC. Quantitative in vivo cell-surface receptor imaging in oncology: kinetic modeling and paired-agent principles from nuclear medicine and optical imaging. Phys Med Biol. 2015;60:R239
Author Biography
 Yu Winston Wang received his Ph.D. degree in Mechanical Engineering from Beihang University, Beijing, China, in 2013. After graduation, he joined Dr. Jonathan Liu's research group as a postdoctoral associate to develop SERS nanoparticles and spectral imaging systems for visualizing cancer biomarkers in tissues. His research interests include Biomedical Optics and Molecular Imaging.
Yu Winston Wang received his Ph.D. degree in Mechanical Engineering from Beihang University, Beijing, China, in 2013. After graduation, he joined Dr. Jonathan Liu's research group as a postdoctoral associate to develop SERS nanoparticles and spectral imaging systems for visualizing cancer biomarkers in tissues. His research interests include Biomedical Optics and Molecular Imaging.
 Nicholas P. Reder received a B.S. degree in Biochemistry from University of Michigan in 2008, a M.P.H. degree in Epidemiology from Emory University School of Public Health in 2010, and a M.D. degree with Honors in Research from Loyola Stritch School of Medicine in 2014. Since 2014, he has been working as a resident in Pathology and a postdoctoral researcher in Dr. Jonathan Liu's research group at the University of Washington. His research interests include Biomedical Optics and Molecular Imaging for cancer diagnosis and surgery.
Nicholas P. Reder received a B.S. degree in Biochemistry from University of Michigan in 2008, a M.P.H. degree in Epidemiology from Emory University School of Public Health in 2010, and a M.D. degree with Honors in Research from Loyola Stritch School of Medicine in 2014. Since 2014, he has been working as a resident in Pathology and a postdoctoral researcher in Dr. Jonathan Liu's research group at the University of Washington. His research interests include Biomedical Optics and Molecular Imaging for cancer diagnosis and surgery.
 Soyoung Kang received a B.S. degree in Biomedical Engineering from University of Southern California in 2010 and a M.S. degree in Medical Systems Engineering from University of Magdeburg, Germany in 2014. She is currently pursuing a PhD degree in the Mechanical Engineering Department at the University of Washington. Her work aims to develop a Raman-imaging system for visualizing cancer biomarkers intraoperatively during tumor resection procedures.
Soyoung Kang received a B.S. degree in Biomedical Engineering from University of Southern California in 2010 and a M.S. degree in Medical Systems Engineering from University of Magdeburg, Germany in 2014. She is currently pursuing a PhD degree in the Mechanical Engineering Department at the University of Washington. Her work aims to develop a Raman-imaging system for visualizing cancer biomarkers intraoperatively during tumor resection procedures.
 Adam K. Glaser received his Ph.D. degree from the Thayer School of Engineering at Dartmouth College in 2015 under the guidance of Dr. Brian Pogue. He is now a post-doctoral research associate in Dr. Jonathan Liu's research group at the University of Washington, working on the development of light-sheet microscopes for slide-free non-destructive pathology of human tissues. In 2017, he was awarded a F32 NRSA postdoctoral fellowship from the NIH/NCI. His research interests include biomedical optics, molecular imaging, and medical physics.
Adam K. Glaser received his Ph.D. degree from the Thayer School of Engineering at Dartmouth College in 2015 under the guidance of Dr. Brian Pogue. He is now a post-doctoral research associate in Dr. Jonathan Liu's research group at the University of Washington, working on the development of light-sheet microscopes for slide-free non-destructive pathology of human tissues. In 2017, he was awarded a F32 NRSA postdoctoral fellowship from the NIH/NCI. His research interests include biomedical optics, molecular imaging, and medical physics.
 Jonathan T.C. Liu received degrees in Mechanical Engineering at Princeton (B.S.E., 1999) and Stanford (M.S., 2000 & Ph.D., 2005). He was a postdoctoral fellow in the Department of Electrical Engineering (Ginzton Labs) and the Molecular Imaging Program at Stanford (2005-2009), and was later appointed as an instructor within the Stanford University School of Medicine (2009-2010). From 2010 to 2014, Jonathan was an assistant professor in the Biomedical Engineering Department at SUNY Stony Brook. In 2014, he was appointed as an assistant professor and then an associate professor (in 2016) in the Mechanical Engineering Department at the University of Washington in Seattle. His laboratory for molecular biophotonics develops optical strategies for improving the diagnosis and treatment of diseases. https://www.me.washington.edu/people/faculty/jonathan_liu
Jonathan T.C. Liu received degrees in Mechanical Engineering at Princeton (B.S.E., 1999) and Stanford (M.S., 2000 & Ph.D., 2005). He was a postdoctoral fellow in the Department of Electrical Engineering (Ginzton Labs) and the Molecular Imaging Program at Stanford (2005-2009), and was later appointed as an instructor within the Stanford University School of Medicine (2009-2010). From 2010 to 2014, Jonathan was an assistant professor in the Biomedical Engineering Department at SUNY Stony Brook. In 2014, he was appointed as an assistant professor and then an associate professor (in 2016) in the Mechanical Engineering Department at the University of Washington in Seattle. His laboratory for molecular biophotonics develops optical strategies for improving the diagnosis and treatment of diseases. https://www.me.washington.edu/people/faculty/jonathan_liu
![]() Corresponding authors: Yu “Winston” Wang. Email: yuwang2edu. Phone: (206)543-4445. Fax: (206)685-8047; Jonathan T.C. Liu. Email: jonliuedu. Phone: (206)543-5339. Fax: (206)685-8047.
Corresponding authors: Yu “Winston” Wang. Email: yuwang2edu. Phone: (206)543-4445. Fax: (206)685-8047; Jonathan T.C. Liu. Email: jonliuedu. Phone: (206)543-5339. Fax: (206)685-8047.

 Global reach, higher impact
Global reach, higher impact