ISSN: 2206-7418
Nanotheranostics 2018; 2(1):21-41. doi:10.7150/ntno.22091 This issue Cite
Review
Nanotechnology-Based Strategies for Early Cancer Diagnosis Using Circulating Tumor Cells as a Liquid Biopsy
1. Key Laboratory of Combinatorial Biosynthesis and Drug Discovery of Ministry of Education, School of Pharmaceutical Sciences, and Brain Center, Zhongnan Hospital, and Medical Research Institute, Wuhan University, Wuhan 430072, China;
2. Taihe Hospital, Hubei University of Medicine, Shiyan, Hubei, 442000, China.
Received 2017-7-26; Accepted 2017-10-10; Published 2018-1-1
Abstract
Circulating tumor cells (CTCs) are cancer cells that shed from a primary tumor and circulate in the bloodstream. As a form of “tumor liquid biopsy”, CTCs provide important information for the mechanistic investigation of cancer metastasis and the measurement of tumor genotype evolution during treatment and disease progression. However, the extremely low abundance of CTCs in the peripheral blood and the heterogeneity of CTCs make their isolation and characterization major technological challenges. Recently, nanotechnologies have been developed for sensitive CTC detection; such technologies will enable better cell and molecular characterization and open up a wide range of clinical applications, including early disease detection and evaluation of treatment response and disease progression. In this review, we summarize the nanotechnology-based strategies for CTC isolation, including representative nanomaterials (such as magnetic nanoparticles, gold nanoparticles, silicon nanopillars, nanowires, nanopillars, carbon nanotubes, dendrimers, quantum dots, and graphene oxide) and microfluidic chip technologies that incorporate nanoroughened surfaces and discuss their key challenges and perspectives in CTC downstream analyses, such as protein expression and genetic mutations that may reflect tumor aggressiveness and patient outcome.
Keywords: nanotechnology, circulating tumor cells, liquid biopsy, in vitro diagnostics.
Introduction
Cancer is one of the leading causes of death in the developed world, primarily due to the lack of effective early detection methods and the prevention of metastasis [1]. Moreover, approximately 90% of cancer-related deaths are due to metastasis [2]. Metastasis occurs when cancer cells detach from the primary tumor or metastatic sites and circulate in the peripheral blood [3-5]. These circulating tumor cells (CTCs) may ultimately invade and colonize surrounding tissue to form a secondary tumor [6]. Since the discovery of CTCs in 1869, researchers have utilized CTCs for the early detection of aggressive cancer and the treatment of advanced disease [7-9]. CTCs are considered a noninvasive “liquid biopsy” of a tumor and are expected to replace surgical tumor biopsy in the monitoring of treatment response and determining the prognosis of patients [10, 11]. Studies have shown that the quantity of CTCs is closely related to disease severity, and CTC count is currently used as a prognostic tool to indicate whether a treatment is effective [12, 13]. Researchers have also analyzed CTCs for certain gene or protein variants that indicate whether the patient's tumor is susceptible to a particular drug [14].
Early diagnosis enables timelier treatment, significantly improves patient outcomes, and is essential for successful therapy [15-17]. The detection of CTCs with high purity and recovery rates has a huge effect on the accurate early diagnosis of cancer and consequently successful cancer treatment. However, CTCs are extremely rare (approximately one CTC is mixed with millions of leukocytes and billions of erythrocytes) in circulating blood, especially, at the early stage of a tumor, making CTC capture a technical challenge [18-20]. Another tremendous challenge is the heterogeneous nature of CTCs, such as differences in their morphology and gene expression, especially during epithelial to mesenchymal transition (EMT)[21]. The rarity and heterogeneity of CTCs in the blood of cancer patients require the development of techniques with high specificity and high sensitivity to find rare tumor cells and to distinguish them from epithelial non-tumor cells and leukocytes. Once detected, CTC enumeration and molecular characterization can be applied to prognosticate cancer classification and predict drug therapy [22, 23]. However, the limited sensitivity of commercially available methods, as well as the complexity and heterogeneity of the disease, limits the widespread acceptance and dissemination of CTC-based diagnostics.
Nanotechnology may be the most promising strategy for achieving an ideal CTC capture device to replace traditional tools. Due to their unique physicochemical properties arising from their high surface area, size, shape, unique optical properties and surface chemistry, nanomaterials (1-100 nm in size, in at least one dimension) are very attractive for cancer diagnosis and therapeutics [24-27]. For CTC enrichment and detection, a key advantage of the use of nanomaterials in cancer detection is their large surface-to-volume ratio compared to that of bulk materials [28]. In particular, this property enables binding of highly efficient targeting ligands that recognize molecules indicative of cancer, allowing for the high recovery and specificity of CTC isolation, detection and characterization. Furthermore, the presentation of multiple binding ligands to a cancer cell, for example, is very important to solve the problem of CTC heterogeneity and enhance an assay's sensitivity. In addition, it has been reported that nanoroughened surfaces have an increased surface area that facilitates cell adhesion, binding, and reactions [29]. Compared with that of normal blood cells, the adhesion preference of tumor cells to nanostructured surfaces makes nanoroughened surfaces an alternative technique for CTC capture. In short, the use of nanomaterials for CTC detection with high sensitivity, high purity, high throughput, and low cost will facilitate the advancement of biological and clinical cancer research.
In this review, we present and discuss the emerging approaches in CTC enrichment and detection using versatile nanoplatforms with the aim of highlighting the role of nanotechnology in advancing basic and clinical CTC research. Various types of nanostructured substrates have been developed for CTC detection, including gold nanoparticles, magnetic nanoparticles, graphene, carbon nanotubes, quantum dots, upconversion particles, poly(lactic-co-glycolic acid) and dendrimers [30-35]. These nanoplatforms often provide chemical stability, biocompatibility and control of surface chemistry properties for CTC isolation. In this review, we also present another widely used technological development, microfluidics platforms, which were developed for capturing rare cells, including CTCs. Microfluidics mainly rely on their small scale, high-throughput capability and large surface area, which could create numerous new opportunities for in vitro cell sorting and detection [36, 37]. For rare cell detection using microfluidics, this microsystem can precisely control the flow behavior, transportation, and biological interactions in the microchannel environment [38-41]. Inspired by the outstanding properties of nanomaterials for CTC separation, researchers have also developed nanostructure-embedded substrates by incorporating nanotechnologies into microfluidics devices to improve CTC separation efficiency and purity [42]. In this review, we only present representative nanotechnologies used for CTC isolation and highlight the aspects of these technologies that enable downstream analyses of CTCs beyond enumeration.
Currently available CTC isolation technologies
Certainly, CTCs are an exciting potential strategy for cancer diagnosis and treatment. Therefore, researchers have focused on new methods for the detection of CTCs to explore the biology of metastases and predict tumor recurrence [43]. Currently, numerous technologies have been developed to isolate CTCs from highly abundant leukocytes and erythrocytes based on their biological and/or physical properties. Physical-based separations are dependent on size [44], deformability [45], density [46], or the dielectric properties of CTCs that are distinct from those of normal blood cells [47]. The physical properties that allow for separation without labeling are summarized as follows: (1) Isolation of CTCs by Ficoll density gradient centrifugation [46]. (2) Selection of CTCs by using the unique differences between cancer cells and blood cells in terms of size (larger) and/or deformability (harder) [48]. Filtration methods generally achieve high-throughput cell isolation. For example, a microfabricated parylene membrane filter (8-μm pore size) could process 7.5 ml of blood within 2 min [49]. The challenges with filtration are low CTC recovery and purity because they are similar in size to leukocytes; thus, the pores of the filters will retain large blood cells while failing to collect small CTCs. (3) Separation of CTCs using hydrodynamic forces based on size and deformability and without the use of any physical structures [50]. This strategy can reduce the risk of device clogging and improve purity. (4) Dielectrophoretic (DEP) cell separation techniques, which can be performed based on differences in cell size and membrane properties [51], allowing the separation of CTCs based on different responses to DEP. The key advantage of physical-based separation platforms is the ability to separate CTCs quickly and economically. However, the overlap in the size/density of leukocytes and CTCs limits the application of these techniques because there is no absolute difference in the physical properties between tumor and non-tumor cells. Thus, such techniques have inherent recovery/purification trade-offs. The biological properties used for CTC isolation primarily rely on the expression of proteins or genes that are not expressed in other blood components. Epithelial cell adhesion molecule (EpCAM) is frequently used as a target for CTC enrichment because it is widely expressed on the cell surface of cancer-derived CTCs and is not detected on normal blood cells [52]. Anti-EpCAM-based immune-identification remains the most commonly used CTC capture platform for the majority of epithelial origin solid tumors.
CellSearch is the only system approved by the US Food and Drug Administration (FDA) for the detection and enumeration of CTCs for clinical use. The system can detect a single CTC in 7.5 ml of blood and has been successfully used to predict patient survival after treatment in a multicenter study [53]. This system uses magnetic nanoparticles functionalized with antibodies that are specific to cell surface antigens such as EpCAM to pull CTCs out from peripheral blood. Isolated cells are subsequently immunostained with fluorescently labeled antibodies and then counted using automated cell image capture and analysis. The CellSearch system has been utilized as a diagnostic and prognostic test and is used to monitor CTCs in patients with various types of cancer, such as prostate, breast, colorectal, pancreatic, gastrointestinal and small lung cancer [54-56]. However, the CTC detection sensitivity of this approach is highly dependent on epithelial markers and is therefore limited by the lack of EpCAM expression in various cancer cells. In addition, CTCs are thought to undergo EMT, in which cells lose epithelial marker expression [57]. Due to the poor stability and heterogeneity of EpCAM expression in cancer cells, particularly with respect to EMT, anti-EpCAM-based CTC capture strategies can be constrained, especially in a low-resource environment. This limitation can be solved by replacing anti-EpCAM with other capture agents such as aptamers, which are generated by a process termed SELEX (Systematic Evolution of Ligands by EXponential Enrichment) [58]. Aptamers are screened oligonucleotide ligands (single-stranded DNA or RNA molecules) with specific affinity for their target, comparable to antibody-antigen interactions, and have demonstrated potential as an alternative capture agent [59]. Aptamers also show properties of high stability, negligible toxicity and immunogenicity, reversible denaturation, and resistance to harsh conditions (detergents, chaotropic salts, organic solvents and pH) [60]. Notably, aptamers can directly target the binding sites, ranging from small compounds to large cell membrane proteins, without requiring prior knowledge of these target molecules.
CTC isolation is particularly challenging due to the issues of CTC loss and low purity. However, none of the abovementioned CTC isolation methods are ideal platforms to meet the application requirements. Most of these isolation strategies include complex processes such as centrifugation, erythrocyte lysis, and washing, which can result in adequate CTC separation. In general, CTC detection and counting is still not part of routine tumor staging in clinical practice because CTCs are extremely rare, less sensitive and specific. As CTC count corresponds to the patient's prognosis, CTC detection techniques should be urgently developed with high efficiency and sensitivity in order to contribute to downstream CTC characterization [61]. At present, nanotechnology has made excellent contributions to the field of biomedical applications, and many researchers take advantage of nanotechnology to improve the selectivity and sensitivity of CTC isolation and to speed up the detection rate [2]. The uniquely appealing physical properties of nanomaterials facilitate their application in CTC isolation and can be used to overcome the limitations of traditional CTC detection methods.
Nanomaterial-based biological CTC detection
Currently, nanotechnology is extensively applied in biomedicine, and numerous researchers have made use of the superiority of nanotechnology to improve the efficiency and sensitivity of CTC detection for the early diagnosis of cancer [62]. Nanostructured substrates have been reported to enhance local topographical interactions between substrates and targeted cell surfaces compared with flat substrates, which enhances cell capture affinity [63]. In addition, CTCs prefer nanostructured surfaces due to the similarity of the cell surface structure [28]. Different types of nanomaterials, such as nanostructured surfaces, quantum dots (QDs), magnetic nanoparticles (MNPs), gold nanoparticles (AuNPs), and polymers, which facilitate cellular attachment, have been reported for CTC detection. These nanomaterials have been demonstrated to improve the specificity and sensitivity of CTC capture devices and are expected to have a potential impact on cancer diagnosis and prognosis.
Nanostructured surfaces for CTC separation
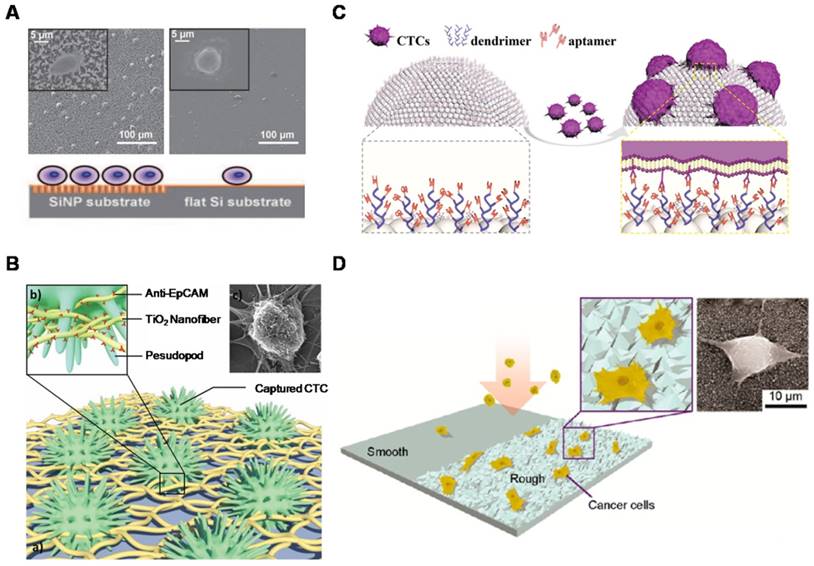
An enormous number of studies have reported that cell pseudopodia can form on nanostructured surfaces, allowing enhanced local topographical interactions between nanostructured substrates and cancer cells compared with unstructured substrates [64, 65]. Wang et al. (Figure 2A) created a 3D nanostructured silicon nanopillar (SiNP) substrate by using a silver and hydrofluoric acid etching process [66]. After being coupled with anti-EpCAM antibody, the modified SiNP substrate provided a higher capture yield (45-65%) than the flat Si substrate (4-14%). The optimized length (longer than 6 μm) and diameter (ranging from 100-200 nm) of SiNPs corresponded to the dimensions of extracellular structures (e.g., microvilli and filopodia). In contrast to the vertical oriented nanopillars described above, Zhang et al. (Figure 2B) fabricated horizontally oriented titanium nanofibers (TiNFs) for CTC detection [67]. The TiNF substrate was simply fabricated by electrospinning the titanium n-butoxide (TBT)/polyvinyl pyrrolidone (PVP) components onto silicon substrates. The device exhibited a capture efficiency of 40-70% when applied to separate tumor cells from a whole blood sample. Due to the advantages, such as precisely controllable dimensions and packing density, biocompatibility, and feasibility of exploring the use of new materials, the electrospun TiNF-deposited substrate was expected to have potential applications for isolation of rare cells, early diagnosis of cancer and molecular biological analysis. These results demonstrated that the synergy benefits between the capture agent (e.g., anti-EpCAM) and nanostructured surfaces resulted in enhanced cancer cell capture. Instead of using anti-EpCAM antibody for CTC capture, Zheng et al. (Figure 2C) developed an aptamer-functionalized barcode-particle technology for the capture and release of different types of CTCs [68]. This device utilized the different characteristic reflection peaks of barcode particles, which could be modified with different types of aptamers, realizing the fluorescence identification of different CTC types. The surface of barcode particles was etched into a nanopatterned topography and decorated with highly branched dendrimer-amplified aptamers, resulting in significantly increased CTC isolation efficiency. Moreover, Chen et al. (Figure 2D) reported a simple and effective platform to capture CTCs by utilizing only the differential adhesion preference to nanorough surfaces of cancer cells and normal blood cells, regardless of their biological and physical properties [29]. The reactive ion etching (RIE)-generated nanorough glass surface that they constructed successfully captured multiple types of tumor cells in the absence of capture antibodies. The rougher the substrate, the more cells were captured.
Immunomagnetic separation of CTCs
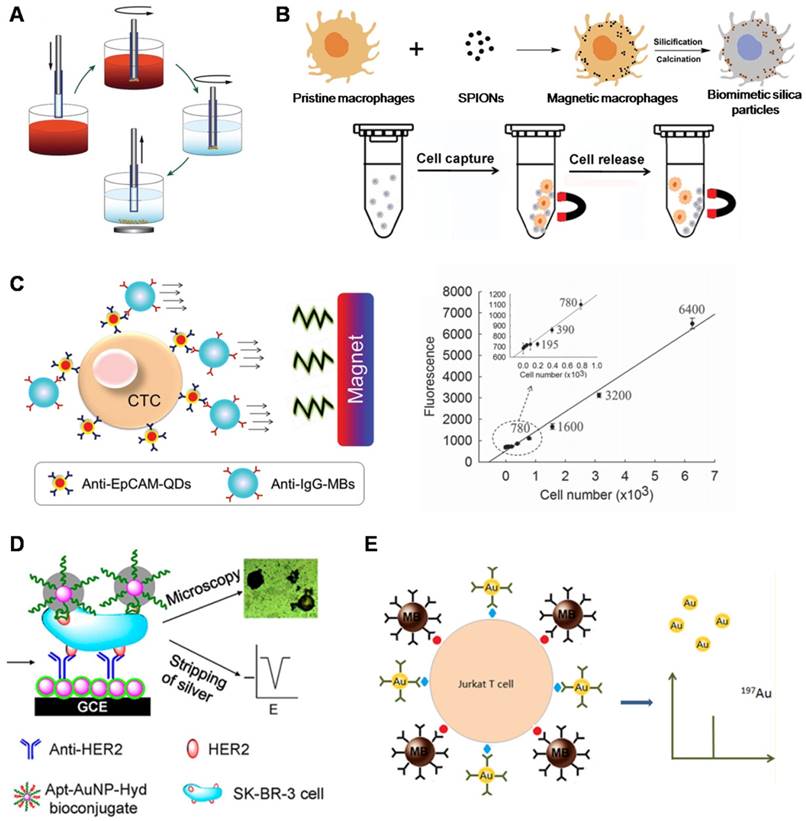
In the rapidly developing nanobiotechnology field, magnetic nanoparticles (MNPs) are one of the most well-established nanomaterials utilized to bind to cells for in vitro separation under an external magnetic field [69]. Iron oxide MNPs, such as magnetite (Fe3O4), due to chemically stable and biocompatible properties, have been most frequently applied in biomedical applications. Immunomagnetic technologies are often performed using anti-EpCAM antibody-functionalized MNPs to specifically target EpCAM-expressing cells. MNP-bound cells are then isolated under an external magnetic field with good sensitivity. Numerous types of immunomagnetic technologies for CTC isolation have been rapidly developed due to their ease of operation and demonstration of high capture specificity and efficiency. Conventional magnetic-activated cell sorting (MACS) systems are employed by loading MNP-treated blood samples into centrifuge tubes [70]. Then, MNP-labeled cells are attracted to the tube wall under an external magnetic field, and unlabeled cells are eluted. These conventional MACS systems have the advantages of simplicity, low cost, convenience, and power function, and they are also capable of separating a large number of cells. Separating rare cells with conventional MACS systems is usually limited by the low sensitivity and capture efficiency because the system is operated manually, and the magnetic field is erratic. MagSweeper is a novel magnetic cell sorting system developed by Powell et al. (Figure 3A) that uses sheath-covered magnetic rods to sweep through the capture wells and magnetically attract magnetic particle-labeled target cells [71]. The device successfully purified CTCs from 70% of primary and metastatic breast cancer patients. The MagSweeper can gently extract high-purity CTCs from normal blood and facilitated robust analysis of single CTCs, providing early insight into CTC heterogeneity. The conventional MACS systems used for cell capture only depend on antigen-antibody interactions. Huang et al. (Figure 3B) developed a “live template” strategy that employed living organisms (e.g., macrophages) incubated with MNPs for magnetic CTC separation [72]. These multifunctional particles have intact cellular topographical features and exhibit enhanced interactions with cell filopodia, resulting in enhanced cell capture efficiency compared with conventional MACS systems.
Electrodeposited nanowires have been used in efficient magnetic cell separation due to their advantages of tunable morphology and composition and their large magnetic moments. Hultgren et al. developed nickel nanowires (35 μm in length, 350 nm in diameter) for cell separation; these nanowires were fabricated by electrochemical deposition into nanoporous templates [73]. The authors showed that nanowires had a higher purity (90%) and yield (49%) than commercially available beads, which might be due to their large surface area and larger magnetic moments. In other studies, they examined the effect of wire length on cell separation efficiency [74, 75], observing that the nanowires in the large size range (5-35 μm in length) achieved high-purity cell separation. They also found that the separation yield increased when the length of the nanowires matched the average diameter of the cells. Compared with commercially available magnetic beads, the Ni nanowires showed a 4-fold increase in cell separation with a purity of 80% and a yield of 85%. Similarly, Hong et al. developed a new method for the ultrasensitive separation and detection of CTC by using multifunctional magnetic nanowires (NWs) for different types of cancer [76]. The electrochemically deposited polypyrrole NWs (16 μm in length) were doped with a high density of 10-nm magnetic nanoparticles and five different types of antibodies (EpCAM, EGFR, N-cadherin, TROP-2 and vimentin) for immune-magnetic selection of CTCs. The elongated structure of the NWs can enhance the cell-NW attachment and increase the number of multivalent binding sites for efficient CTC isolation. The NWs could capture and recognize CTCs from the blood of breast cancer patients, especially from 29 cases of non-metastatic early cancers. This multifunctional magnetic NW-based CTC capture method may provide an effective tool for cancer prognosis, early cancer diagnosis and treatment response assessment.
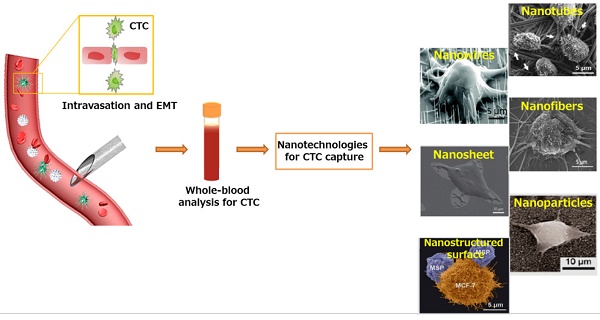
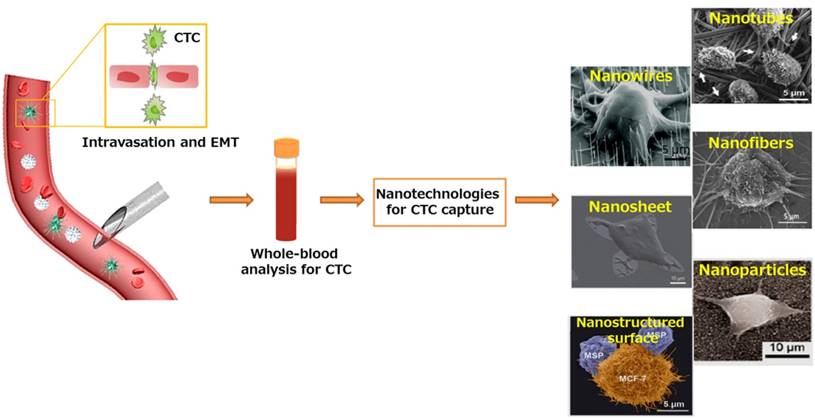
Metastatic CTC as a liquid biopsy for early cancer diagnosis. Nanotechnologies developed for sensitive CTC isolation. SEM images of CTCs captured on nanorough substrates with different morphologies. Images of Nanowires: Copyright IOPscience, 2012. Reproduced with permission from reference [166]. Images of Nanosheet: Copyright Nature, 2013. Reproduced with permission from reference [31]. Images of Nanofibers and Nanostructured surface: Copyright Wiley, 2012 and 2015. Reproduced with permission from references [67] and [72]. Images of Nanotubes: Copyright Elsevier 2006. Reproduced with permission from reference [167]. Images of Nanoparticles: Copyright American Chemical Society, 2012. Reproduced with permission from reference [29].
Nanostructured surfaces for CTC enrichment. (A) 3D silicon nanopillar (SiNP) substrates showing significantly enhanced cell binding affinity in contrast to flat substrates. (B) Horizontally packed TiO2 nanofibers (TiNFs) for improved CTC capture by combining anti-EpCAM antibody and cancer cell-preferred nanoscale topography. SEM image of a target cell captured on TiNF substrate (right), with fully outspread pseudopod attached to the surface of the TiNF. (C) Aptamer-functionalized barcode particles for CTC isolation. Dendrimer is decorated on the surface of the barcode particles for enhanced CTC capture. (D) Cancer cells prefer to adhere to ion-etched glass surfaces. Insets are zoom-in (left) and SEM (right) images of cancer cells captured on nanorough glass surfaces. (A) Copyright Wiley, 2009. Reproduced with permission from reference [66]; (B) Copyright Wiley, 2012. Reproduced with permission from reference [67]; (C) Copyright Wiley, 2014. Reproduced with permission from reference [68]; (D) Copyright American Chemical Society, 2012. Reproduced with permission from reference [29].
Quantum dots for CTC capture and fluorescent detection
Quantum dots (QDs) are inherently fluorescent nanoparticles with high quantum yields and tunable emission wavelengths that are desirable for biological imaging applications and for the development of novel cancer diagnostics [77]. QDs exhibit longer fluorescence lifetimes compared with organic fluorophores [78]. This property is especially significant in the application of QDs for enhancing the sensitivity of surface marker-dependent CTC capture and sorting, particularly when CTCs may be in low abundance at the early stages of the disease. The size dependence of the absorption and emission wavelengths of QDs enables the tunable design of QDs for a range of imaging applications, especially in multicolor labeling for the simultaneous detection of multiple targets [79]. In addition, size-tuned QDs with different emission wavelengths corresponding to the same excitation wavelength can be applied to capture and sort heterogeneous CTCs [80]. To develop a specific and sensitive diagnostic platform, Xie et al. constructed a novel multifunctional nanobioprobe for CTC detection and release [81]. After assembling alginate onto the surface of QDs with Ca2+ activation, they immobilized biotinylated anti-EpCAM to fabricate a multifunctional nanoprobe that had the advantage of specific cell capture and EDTA-assisted cell release with 86% and 65% efficiency, respectively. Similarly, Min et al. (Figure 3C) employed primary anti-EpCAM-modified QDs to target and enumerate CTCs and secondary anti-IgG-magnetic beads (MBs) to isolate the attached cells [33]. They demonstrated that the fluorescence intensity of QDs was consistent with the number of cells captured. Compared to previous work based on the use of only QDs or MBs, this platform shows prominent synergy benefits between nanoparticles and MBs with high capture efficiency and a simple CTC quantification method.
Functional nanomaterials used for CTC enrichment. (A) MagSweeper device showing magnetic rods sheathed in plastic for CTC capture and release. (B) Preparation of the “live template” strategy to produce particles with magnetic and topographic properties. Cells were captured and released by an external magnetic field. (C) CTC capture using QDs and MNPs. Quantification of captured CTCs as a function of the fluorescence intensity of the anti-EpCAM-QDs (right). (D) Electrochemical immunosensor based on a hydrazine-AuNP-aptamer for the detection of the HER2 protein and HER2-overexpressing SK-BR-3 cells. The silver-stained target cells were visualized under microscopy and quantitatively analyzed using stripping voltammetry. (E) Immunomagnetic separation of Jurkat T cells with MBs. The captured cell was then bound to AuNPs for inductively coupled plasma mass spectrometry (ICP-MS) measurement. (A) Copyright Public Library of Science (PLoS), 2012. Reproduced with permission from reference [71]; (B) Copyright Wiley, 2015. Reproduced with permission from reference [72]; (C) Copyright Wiley, 2015. Reproduced with permission from reference [33]; (D) Copyright American Chemical Society, 2012. Reproduced with permission from reference [85]; (E) Copyright American Chemical Society, 2014. Reproduced with permission from reference [87].
Gold nanoparticles for CTC capture and optical detection
Gold nanoparticles (AuNPs) have emerged as a unique nanoplatform for CTC detection owing to their advantages of simple synthesis, unique spectral properties, and multiple surface functionalities [82, 83]. The thiolated aptamer can be self-assembled to the AuNP surface through S-AuNP bonds [30]. The ease of assembly of AuNPs with multiple aptamers provides a versatile platform for efficient cell capture. The tunable optical properties of AuNPs have been used to detect CTCs because the binding between gold nanoparticles and CTCs can be measured quantitatively using a surface plasmon resonance shift or a photoacoustic signal [84]. To improve the detection sensitivity at low target concentrations, a signal amplification method was developed by employing self-assembled AuNPs for electrochemical diagnosis of breast cancer (Figure 3D) [85]. HER2-overexpressing cells were initially captured by anti-HER2-conjugated AuNPs, and the number of cells was determined by the silver metal deposited on AuNPs through square wave stripping voltammetry. This method exhibited highly sensitive detection of SK-BR-3 breast cancer cells from human blood samples with a detection limit of 26 cells/ml. In a similar study, Yi et al. provided an electrochemical method for the sensitive detection of tumor cells [86]. Briefly, cancer cell capture was first accomplished using aptamer-conjugated-AuNPs. As a detection probe, silver was deposited onto the AuNPs, allowing for simple amplification of sensitive electrochemical detection and achieving cell detection with high sensitivity (as few as 10 cells). By combining magnetic immunoassay with AuNP immunoprobes, Zhang et al. (Figure 3E) designed a novel immunoassay for the efficient and fast separation of tumor cells [87]. Magnetic nanobead probes were used for the capture of targeted cells, and AuNPs were used as detection probes for the sensitive and precise detection of cells via inductively coupled plasma mass spectrometry (ICP-MS).
Other nanomaterials for CTC detection
Carbon nanotubes (CNT) have outstanding electronic properties and have been used in the electronic detection of tumor cells from whole blood samples [32]. Liu et al. developed a sensitive CNT-based biosensor for cancer cell detection by taking advantage of the good conductivity of multi-walled CNTs (MWCNTs) [88]. The biosensing mechanism relies on the binding of anti-EpCAM antibodies to tumor cells, which results in increased electron transfer resistance. The authors showed that the electrical response was linear with the logarithm of the concentration of tumor cells, with a detection limit of 5 cells per ml of blood. Upconversion NPs (UCNPs) exhibit an anti-Stokes shift and show strong emission under NIR excitation. UCNPs capacitate ultrasensitive imaging in biological samples due to the minimization of spontaneous fluorescence from cells and tissues. Fang et al. developed for the first time the use of UCNPs as nanoprobes for fluorescence-based CTC detection in combination with immunomagnetic enrichment [34]. They used aptamer-coated NaYF4 (Yb:Er) UCNPs that targeted cancer cells and demonstrated that the fluorescence intensity was linearly correlated with the concentration of captured cancer cells, with a detection limit of 10 cancer cells in 10 ml of blood.
Microchip-based biological CTC detection
Compared with conventional methods requiring multiple steps to sort, enumerate, and analyze CTCs, which potentially lead to CTC loss, microchip-based technologies provide excellent platforms to perform the isolation, transport, and analysis of rare cells in one chip [89]. Microfluidic platforms have become the mainstream of CTC isolation and detection methods due to their numerous advantages, including low cost, small sample-volume requirements and the ability to integrate with other techniques to improve the efficiency of the device [90]. To improve the isolation efficiency, enhancing the surface interactions between the cells and the surface of the channels is important. However, the laminar flow that causes the cell to move along a straight line parallel to the fluid channels is more dominant in microfluidic channels.
Due to the large surface-to-volume ratio, integrated nanostructures can be linked with ligands (e.g., nucleic acid aptamers or antibodies) with much higher densities, facilitating surface-dependent applications, such as cell capture based on affinity. For example, most of the efforts to increase the sensitivity of cell capture have relied on integrating complicated structures inside the microfluidic devices, such as silicon nanopillars, sinusoidal channels, and microposts [42, 91, 92], to enhance cell-ligand interactions. In addition, when nanostructures are integrated into microfluidic devices, they reduce the rate of cells rolling in the channels, thereby further enhancing cell affinity. For example, microfluidic combination systems, such as chaotic mixers, can greatly improve the capture efficiency and purity of CTCs [91]. In brief, the integration of nanostructures into microfluidic devices can be a strategic method for CTC isolation due to the synergistic effects of combining nanotechnology and microfluidic technology. For example, in 2007, Nagrath et al. developed a microfluidic cell capture platform called the “CTC Chip” with sensitivity superior to that of the FDA-approved CellSearch platform [93]. The CTC Chip was fabricated with 78,000 antibody (EpCAM)-functionalized microposts (100 μm tall, 100 μm in diameter and 50 μm gaps between microposts) to enhance cell-antibody interactions compared to simple flat microfluidic channels. With optimized shear force and flow velocity, this device successfully identified CTCs in 115 of 116 samples from colon, prostate, lung, breast, and pancreatic cancer patients with approximately 50% purity.
Nanostructure-embedded microchips for CTC detection. (A) Enhanced cell capture using multiple aptamer-modified AuNPs for multivalent interactions. (B) A chaotic mixing chip induces enhanced interactions between anti-EpCAM-modified SiNP substrates and flowing cells. (C) A graphene oxide (GO) chip for sensitive CTC capture. GO nanosheets are adsorbed onto the gold pattern. PEG-functionalized GO nanosheets modified with anti-EpCAM antibodies for CTC targeting. (A) Copyright American Chemical Society, 2013. Reproduced with permission from reference [94]; (B) Copyright Wiley, 2011. Reproduced with permission from reference [91]; (C) Copyright Nature, 2013. Reproduced with permission from reference [31].
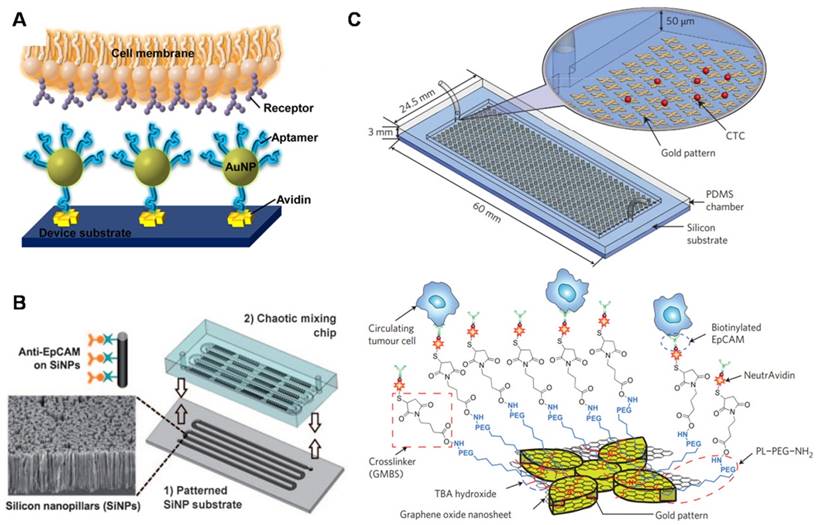
Nanostructure-embedded microchips for CTC detection
Numerous microchip-based CTC detection technologies have been optimized by incorporating nanomaterials into microfluidics to facilitate CTC isolation and characterization [42]. Finely patterned (deposited or etched into nanowires, nanoparticles, or nanopillars) Au or silicon, for example, can be incorporated into a microfluidic chip to increase the surface contact with extracellular structures. Sheng et al. (Figure 4A) designed an efficient CTC isolation platform by depositing AuNPs onto a microfluidic channel and assembling a number of aptamers [94]. Increased cellular binding (39-fold) was achieved compared with that obtained using aptamer-coated flat surfaces, and the capture yield increased from 49 to 92%, indicating great potential for sensitive CTC isolation. Stott et al. introduced a herringbone chip (HB-chip), a second generation CTC Chip, using herringbone microstructures to disrupt the laminar flow streamlines, thereby improving the cell-antibody interactions and achieving a higher cell recovery yield (26.3% improvement under a flow rate of 1 ml/h) than the CTC Chip[95]. Whereas the HB-chip simply increased the likelihood of cell-antibody interactions, Wang et al. (Figure 4B) patterned an HB-chip with nanostructured silicon substrates to enhance the interactions between the nanoscale components of the cellular surface (e.g., microvilli and filopodia) and the anti-EpCAM-conjugated SiNP array [91]. The resulting synergistic effects led to a high CTC capture performance, and the recovery rate of cancer cells in artificial samples was as high as 95%. However, although the herringbone-structured devices have been widely used for CTC isolation, it is still uncertain whether enhanced flow mixing is necessary to increase cell surface interactions. Liu's group investigated the geometric effects of herringbone structures on the CTC isolate performance by applying a computational model incorporated with adhesion probability [96]. This developed model can be used to optimize the parameters of microfluidics for cell capture. For example, they demonstrated that increased channel width resulted in decreased capture efficiency and that the number of grooves did not significantly affect the capture efficiency. To extend the herringbone-structured devices, Liu's group developed a wavy-herringbone (wavy-HB)-structured microfluidic device for efficient and selective isolation and release of CTCs from whole blood samples [97]. The cells captured on anti-EpCAM-coated magnetic particles (MPs) were simply and efficiently trapped in the HB structured microfluidic device by an external magnetic field and then released from the device after removal of the magnetic field. The authors demonstrated that a higher yield was obtained using the wavy-HB structured device (92%) than using a groove-HB patterned device (48%) and a flat device (21%).
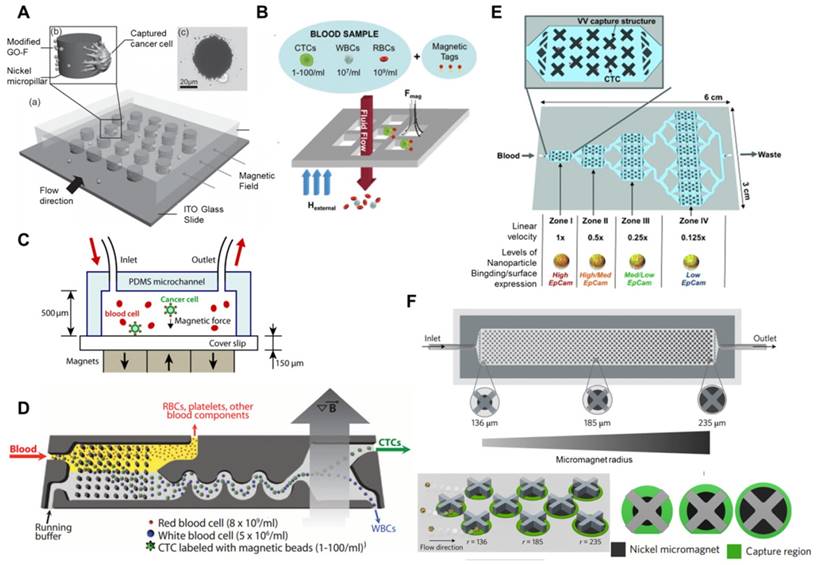
Microchip-based immunomagnetic CTC enrichment. (A) A micropillar device with modified GO-coated Fe3O4 MNPs (GO-F). Image of a cancer cell captured on a nickel micropillar (upper). (B) A magnetic sifter device used for CTC capture from whole blood samples. Magnetic tag-labeled CTCs were captured at the pore edges and unlabeled cells passed through the pores under fluid flow. (C) Microchip design for immunomagnetic detection of Fe3O4 magnetic nanoparticle-labeled cancer cells. (D) CTC-iChip showing the positive selection mode. Whole blood is premixed with immunomagnetic beads, and magnetically labeled CTCs are preferentially forced into a separate outlet. (E) A multizone velocity valley device for isolating magnetically labeled CTCs in four different regions of varying linear velocities. Cells with high EpCAM expression were trapped in zone I, and cells with medium-to-low EpCAM levels were trapped in later zones. (F) The MagRC approach for separating and in-line profiling of heterogeneous CTCs. The chip contains 100 distinct zones with varied magnetic field strengths. X-shaped structures generate local regions of low velocity and circular nickel micromagnets enhance the externally applied magnetic field. Cells with high surface marker expression levels are captured in the earliest zones, and those with low surface marker expression are captured in the later zone of the chip. (A) Copyright Wiley, 2011. Reproduced with permission from reference [92]; (B) Copyright Royal Society of Chemistry, 2013. Reproduced with permission from reference [99]; (C) Copyright Royal Society of Chemistry, 2011. Reproduced with permission from reference [100]; (D) Copyright The American Association for the Advancement of Science, 2013. Reproduced with permission from reference [102]; (E) Copyright Wiley, 2015. Reproduced with permission from reference [103]; (F) Copyright Nature, 2017. Reproduced with permission from reference [104].
The unique optical properties and ease of surface modification make graphene oxide an attractive material for CTC binding and detection applications. For example, Yoon et al. (Figure 4C) developed a sensitive CTC capture method using anti-EpCAM-coated graphene oxide (GO) nanosheets that were prepared on a silicon substrate with flower-shaped gold patterns [31]. The recovery rates reached 73% when samples were spiked with a low concentration (3-5 cells per ml blood) of breast cancer cells. Without the GO sheet, the chip only isolated 13.3% of spiked cells. The GO chip was able to isolate CTCs from metastatic cancer patients and early-stage cancer patients. Both HER2+ and HER2- cells were isolated from breast cancer cells, enabling downstream analysis of the heterogeneity of cancer cells. By combining the high sensitivity of GO for CTC detection with the high efficiency of MNPs for CTC magnetic separation, Yu et al. (Figure 5A) developed a nickel micropillar device assembled with GO-coated MNPs for the capture and controllable release of cancer cells induced by an external magnetic field [92]. When the magnetic field was removed, 92.9% of the captured cancer cells were released, 78% of which were viable. Other nanomaterials, such as halloysite nanotubes, have also been employed for CTC detection. Halloysite nanotubes are typically 500 nm to 1.2 μm in length and 40-200 nm in diameter, which are uncharacteristically large dimensions for nanoparticles. The average rolling velocity of cells with the nanotube-coated device was demonstrated to be greatly reduced compared to the control device, resulting in enhanced call adhesion. The halloysite nanoparticle coating was also found to increase the surface area, binding more target molecules and thus achieving significantly enhanced target cells capture.
Microchip-based immunomagnetic assay
Similar to the conventional MACS system, microchip-based immunomagnetic assays are also commonly used devices for CTC isolation due to the combined benefits of the microfluidic device and immunomagnetic separation [98]. In microchip-based immunomagnetic assays, CTCs are either fixed and captured on the substrate or magnetically driven to different streamlines and collected at designated outlets. For example, Earhart et al. (Figure 5B) recently developed a magnetic sifter for CTC capture and release with high-throughput [99]. In the presence of magnets, MNP-labeled cells were trapped at the edge of the 40 μm pores in a silicon nitride membrane, while unlabeled cells were removed by the flow. Release occurred by removing the external magnet followed by a buffer wash, and an average of 92.7% release efficiency was achieved. In addition to enumeration, this device was also capable of detecting gene mutations in lung cancer patients by using mutation-specific antibodies, which is important for selection of appropriate therapy approaches. In another study, researchers (Figure 5C) developed a high-throughput (optimal flow rate of 10 ml/h) microchip-based CTC immunomagnetic separation method [100]. The device was simply fabricated by laying permanent magnets under the chip, and cancer cells labeled with anti-EpCAM-coated MNPs were deposited at the bottom wall of the glass coverslip by the magnetic field. Due to the risk of CTC loss using positive selection methods, Chen et al. described a negative enrichment approach, using a disk-based microchip for the immunomagnetic negative separation of rare cells [101]. By utilizing multiple concentric-circular magnets, they were able to capture magnetically labeled non-target cells and achieve a cell capture yield of 60%.
Conventional MACS platforms have had great success in CTC isolation and cancer diagnosis. Due to the cell heterogeneity, new technology is required to obtain controllable hydrodynamic forces and magnetic fields acting on targeted cells to improve separation. The CTC-iChip, developed by Ozkumur et al., (Figure 5D) is an automatable CTC sorting technology that combines the strengths of microfluidics and the benefits of magnetic-based cell isolation [102]. CTCs were labeled with anti-EpCAM-coated MBs, and leukocytes were labeled with CD45 and the granulocyte marker CD15. Then, the device utilized a hydrodynamic size-based sorting method to remove smaller cells, and the remaining cells (large leukocytes and cancer cells) were aligned into a single line via inertial focusing. Finally, labeled cancer cells were separated from non-labeled cells under a magnetic field. Isolating CTCs dependent or independent of tumor surface markers would make it possible to diagnose all types of cancers. To separate CTCs with different phenotypes, Mohamadi et al. (Figure 5E) developed a multiscale immunomagnetic approach to trap CTC subpopulations in different compartments of a fluidic chip based on the differential expression of surface markers [103]. CTCs with a higher level of EpCAM expression might be labeled with higher density of MNPs, which require a higher drag force to overcome the low magnetic force, whereas CTCs labeled with fewer MNPs would continue moving and be trapped only when they entered a zone with lower drag force at a lower flow rate. The sensitivity of this device made it capable of sorting heterogeneous CTCs and investigating EMT in patient CTCs. Poudineh et al. (Figure 5F) reported a magnetic ranking cytometry (MagRC) approach to separate and in-line profile heterogeneous CTC phenotypes based on the longitudinal profile of magnetic field gradients [104]. CTCs with high EpCAM expression were captured in the earliest zones of the MagRC Chip, while low EpCAM-expressing cells were captured in the later zones of the chip. They demonstrated that this system was able to profile CTCs accurately, with a detection limit of 10 cells/ml in unprocessed blood, and monitor changes in CTC levels and phenotypes.
Nanotechnology-based CTC release assay
Although the detection and enumeration of CTCs provides significant diagnostic information, CTC-derived downstream molecular characterization may provide more valuable insights into the mechanism of cancer metastasis, accurate diagnosis, and therapeutic choices [105, 106]. To carry out CTC molecular and functional analyses, technologies must be able to not only isolate CTCs with high efficiency and purity but also release CTCs without disrupting CTC viability or function. Immunomagnetic technologies for CTC isolation have the advantage of releasing captured cells simply by removing the magnetic field. However, a drawback of these methods is that the MBs attached onto cells cannot be released, which will affect the cell viability and post-release cell cultures. Accordingly, most cell capture platforms have demonstrated challenging captured cell release or poor cell viability after release. Hou et al. (Figure 6A) pioneered a “NanoVelcro” CTC assay fabricated with chemically etched silicon nanowire (SiNW) substrates, which were covalently grafted with anti-EpCAM-functionalized thermally responsive polymer brushes, poly(N-isopropylacrylamide) (PIPAAm), for CTC capture and release [107]. Due to the thermally responsive PIPAAm switch, the biocompatible polymer can reversibly bind and release CTCs. At 37 ℃, the cells are captured on hydrophobic domains through biotin-streptavidin interaction. When the temperature is reduced to 4 ℃, the backbone of the polymers exhibit a hydrophobic-to-hydrophilic switch, resulting in detachment of the captured cells from the substrates. This platform demonstrated cancer cell capture at 37 ℃ with high efficiency (>70%) and the subsequent release of these cells at 4 ℃ with high efficiency (90%) and viability (90%). Although the polymer nanostructure (e.g., PIPAAm) was reported to release immobilized cells with high viability, more feasible and economical releasing techniques are desired for practical release of captured CTCs. To achieve this goal, Huang et al. reported a self-sacrifiable nanofilm substrate for CTC capture and release [108]. The platform was simply fabricated via in situ self-assembly of MnO2 hollow nanoparticles onto a glass substrate to form a monolayer thin film. The MnO2 thin film was able to be dissolved by low concentrations of oxalic acid, offering the feasibility of releasing the captured cells. The high transparency of the thin film also made it convenient for direct observation and tracking of the captured cells and the subsequent release. Although the self-sacrifiable platform has the advantages of simple fabrication, convenience, and cost-efficiency, it is limited by low cell purity, similar to most static cell capture devices, and low viability (50%) of released cells due to the cytotoxicity of the oxalic acid solution. To achieve high specificity, purity, and especially high viability of the released cells, Huang et al. later used biodegradable gelatin nanomaterial-coated silica beads for facile CTC capture and release [109]. Gelatin is a natural protein extract from animal collagen, with excellent biocompatible properties and biodegradability. The gelatin coating can be degraded by matrix metalloproteinase-9 (MMP-9), which is a type of protease expressed in tumor tissues, making the noninvasive (with a cell viability of more than 90%) release of CTCs more feasible compared with some enzyme-induced cell release strategies. Considering the heterogeneity of EpCAM expression in cancer cells and the difficulty in releasing captured cells from antigen-antibody affinity-based binding, Shen et al. (Figure 6B) developed a next-generation aptamer-coated NanoVelcro chip for efficient CTC capture and release [110]. A surface-grafted aptamer on a SiNW substrate can be specifically cleaved via enzymatic treatment, resulting in specific CTC release and negligible viability (~80%) disruption and allowing the desired molecular and functional analyses of CTCs. In another enzyme-induced CTC release assay, three different types of hybrid nanoparticles (HNPs) consisting of different QDs (Qdot (525), Qdot (565) or Qdot (625)), antibodies (EpCAM, EGFR or HER2) and double-stranded DNA (dsDNA) were utilized for the specific isolation and release of different breast cancer cell subtypes [111]. Qdots that emit different fluorescent signals reflect the different expression of cell surface proteins. The antibodies were used for targeting different types of cancer cells, and the capture yields of MCF-7, SK-BR-3 and MDA-MB-231 were 81.3%, 91.2% and 90.0%, respectively. The dsDNA allowed the capture and release of cells after restriction enzyme treatment, and the average release efficiency of the captured cancer cells was 86.1%. This strategy has the potential to detect multiple types of CTCs and selectively recover the desired cells, which would greatly increase the application of CTCs in mechanistic studies and clinical management.
These CTC release assays have been developed for the noninvasive recovery of isolated CTCs to conduct CTC culture and characterization. Degradable coatings, mainly polymer coatings, have been widely used for CTC capture and release, which is due to their ease of degradation by external operations, such as enzymatic degradation, UV exposure or replacement temperatures. However, these methods are limited by the impact of the release process on cell viability. In addition, these CTC release methods are also hindered by their low purity because the majority of the captured cells that they can release are contaminated by normal blood cells. Nucleic acid aptamers offer several advantages over antibodies in CTC isolation, such as low cost, ease of synthesis, high chemical stability, highly specific molecular recognition and ease of digestion by exonucleases. However, the disadvantages of the aptamer-based CTC isolation methods cannot be overlooked, such as their susceptibility to nuclease degradation when processing blood samples. The greatest drawback of the aptamers might be their specificity for CTCs because the aptamers are usually selected from a large random sequence pool based on cancer cell lines that may differ significantly from CTCs. Microchip-based hydrodynamic separation and in situ culture of CTCs might provide alternative strategies for CTC capture and analysis that do not require an additional release process. However, most CTC release platforms have a certain influence on CTC viability. The development of a novel platform that increases CTC capture efficiency and sensitivity and enables downstream CTC analysis would significantly enhance the application of CTCs in cancer prognostics and diagnostics.
Single-cell isolation and analysis
Most cell capture methods isolate and analyze the bulk of CTCs, neglecting the intercellular heterogeneity between individual CTCs, such as cell heterogeneity in protein and gene expression levels and the response to environmental and chemotherapeutic stimuli [112]. Molecular and gene analysis methods have the advantage of being more sensitive than immunocytochemistry but the disadvantage of low specificity induced by even minor contamination [113]. To avoid the probability of obtaining false positive results due to illegitimate gene amplification in non-tumor cells, a high quality nucleic acid template is required. Currently, single-cell analysis has greatly improved in CTC analysis and has become a significant tool for revealing the biological functions of individual cells [114]. The measurement of molecular signatures within a single cell facilitates the full understanding of the cellular heterogeneity under complex tissue microenvironments. Single-cell analyses have demonstrated that individual cells show marked diversity and heterogeneity, even those within homogeneous tissues [115]. There are several factors that may affect the accuracy of single-cell analyses. The most important factor might be the choice of the CTC isolation platform, which requires the ability to isolate the “real” CTCs before molecular profiling. Single-cell technologies developed for the precise analysis of cancer heterogeneity and subpopulations would allow drug sensitivity testing and better personalized treatment decisions for cancer patients [115]. For example, it has been reported that HER2 overexpression in CTCs from breast cancer patients is a significant determinant of therapy [116]. Furthermore, technological improvements resulting in purer single-CTC isolates would facilitate the early detection of disease and the discovery of biomarkers to monitor disease progress during anticancer therapy.
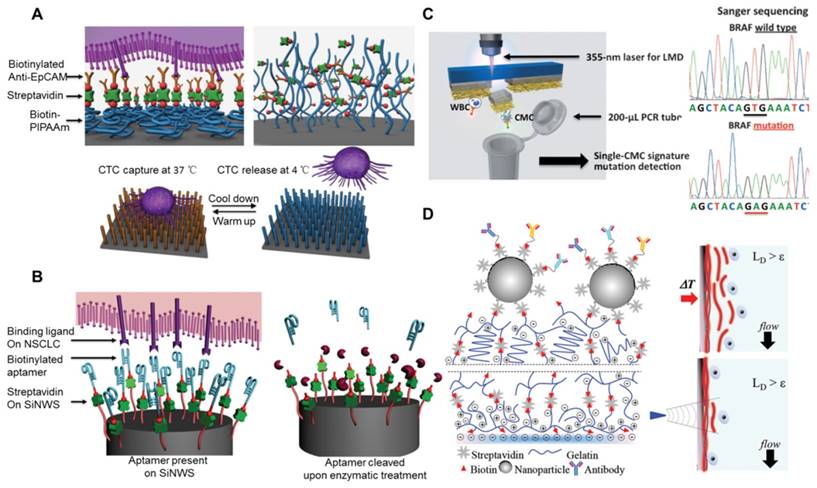
Strategies for CTC release and single-cell analysis. (A) NanoVelcro CTC release assay based on thermal responsive polymer brushes (PIPAAm), which are covalently modified onto a silicon nanowire substrate (SiNWS). (B) Aptamer-coated NanoVelcro Chip for capturing and releasing non-small cell lung cancer (NSCLC) CTCs from blood samples. Exonuclease digestion of DNA aptamers to release CTCs from the aptamer-functionalized SiNWS. (C) Laser microdissection (LMD) technique for single circulating melanoma cell (CMC) isolation. Sanger sequencing of the individually isolated cells. One CMC showed the BRAFV600E mutation and the negative control showed no BRAFV600E mutation. (D) Layer-by-layer gelatin nanocoating for bulk CTC release by raising the temperature to 37 °C or single-cell release of CTCs with a microtip to dissolve localized regions of the nanocoating. (A) Copyright Wiley, 2012. Reproduced with permission from reference [107]; (B) Copyright Wiley, 2013. Reproduced with permission from reference [110]; (C) Copyright Wiley, 2013. Reproduced with permission from reference [120]; (D) Copyright Wiley, 2015. Reproduced with permission from reference [122].
Summary of nanotechnologies for CTC detection.
| Nanomaterial /Device | Description | Affinity ligand | Efficiency % | Purity % | Viability % | Cancer type Metastatic (M) | Ref |
|---|---|---|---|---|---|---|---|
| Magnetic nanoparticles (MNPs) | Ease of surface modification, controllable size, superparamagnetism and response to a magnetic field, high stability of surface chemistry, biocompatibility. | EpCAM | >94 | -- | 90.5 | Colon/liver /lung/breast | [159] |
| -- | >75 | -- | -- | -- | [160] | ||
| EpCAM/EGFR/HER2 | 90 | -- | -- | -- | [161] | ||
| Gold nanoparticles (AuNPs) | Simple synthesis, ease of surface modification, unique spectral properties, thiolated aptamers can be modified on AuNP surfaces through Au-S bonds. | HER2 | -- | -- | -- | Breast | [85] |
| CD2/CD3 | -- | -- | -- | -- | [87] | ||
| Quantun dots (QDs) | Inherent fluorescence, controllable size, long fluorescence lifetime, tunable emission wavelengths. | EpCAM | 86 | -- | 70 | -- | [81] |
| EpCAM | 70-80 | 18-23 | -- | -- | [33] | ||
| Graphene oxides (GO) | High surface area-to-volume ratio, ease of surface modification, controllable size of its sheets, unique optical properties, biocompatibility. | EpCAM | 73 | -- | -- | Breast/lung /pancreatic | [31] |
| >40 | -- | 78 | -- | [92] | |||
| Carbon nanotubes | High surface area-to-volume ratio, good conductivity. | EpCAM | -- | -- | -- | Liver | [88] |
| Nanoroughened surfaces | The preference of cancer cells adhere to nanorough surfaces compared with normal blood cells. | -- | 80 | 14-84 | -- | -- | [29] |
| -- | 90 | -- | -- | -- | [28] | ||
| Si Nanopillars, (SiNP) | Similar size to nanoscale components of the cell surface (e.g., filopodia and microvilli), allowing for increased local topographic interactions. | EpCAM | >40 | -- | 84-91 | -- | [66] |
| EpCAM | >70 | -- | 90 | -- | [107] | ||
| Aptamer | >80 | >95 | 78-83 | NSCCL | [110] | ||
| Nanofibers | Ultralong nanofibers with controllable diameters that have similar dimensions with extracellular matrix (ECM) scaffolds and cell surface components. | EpCAM | 40-70 | -- | -- | Colorectal /gastric | [67] |
| CD146 | 87 | -- | -- | Melanoma | [120] | ||
| Herringbone- Chip (HBCTC-Chip/HB-chip) | A microfluidic mixing device with patterned herringbones on their upper surface to disrupt the laminar flow streamlines that cells travel. The chaotic microvortices increase the interactions between antibody-modified chip surfaces and target CTCs. | EpCAM | 91.8 | 14 | 95 | M-prostate | [95] |
| -- | -- | -- | M-pancreatic | [139] | |||
| -- | -- | -- | Prostate | [141] | |||
| EpCAM/EGFR/HER2 | 80-90 | -- | -- | Breast | [150] | ||
| 75-95.9 | 78-90 | 85-90 | Breast/Lung | [122] | |||
| 80 | 53 | 90 | M-lung | [162] | |||
| -- | -- | -- | Breast/prostate | [3] | |||
| CSPG4/MCAM | >90 | 0.3 | -- | M-melanoma | [127] | ||
| MagSweeper | An immunomagnetic separation technology functionalized with a magnetic rod covered with a plastic sheath. Blood cells will be washed away by the movement of the magnetic rod and magnetic particles attached cells will be captured by a magnetic force produced by the magnetic rod. | EpCAM | >80 | 100 | -- | M-breast | [163] |
| >85 | -- | -- | M-prostate | [164] | |||
| -- | -- | 50 | Breast | [71] | |||
| MagSifter | A magnetic sifter device functionalized with a section of a patterned pore array. Magnetically labeled target cells are captured at the pore edges and unlabeled cells pass through the pores. | EpCAM | >91.4 | -- | -- | Lung | [99] |
| EpCAM | -- | -- | -- | Lung | [14] | ||
| Microchip- based immunomagnetic | A microchip-based immunomagnetic separation that combines an immunomagnetic assay with a microfluidic device. | EpCAM | 86 | -- | -- | -- | [100] |
| EpCAM | 66 | -- | -- | Breast/prostate /lung | [165] | ||
| CTC-iChip | A microchip-based immunomagnetic device performed by positioning cells in a near-single file line and targeted cell can be precisely deflected using a minimal magnetic force. | EpCAM | 96.7 | >0.1 | -- | Prostate/breast /pancreas /colorectal/lung | [102] |
| Magnetic ranking cytometry (MagRC) | A microchip-based immunomagnetic separation with X-shaped structures within the microfluidic channel that generates regions with slow flow and can be accurate for the in-line profiles of CTCs at the single-cell level. | EpCAM | 90 | -- | -- | Prostate | [104] |
Considering that individual cells isolated and captured in the chambers of a microchip can be easily lysed for subsequent molecular analysis [117, 118], microfluidic technology is thought to hold great promise for single-cell analysis of CTCs and detecting viable CTCs with satisfactory efficiency and purity[119]. Park et al. utilized the MagSifter system for the high-throughput magnetic separation of CTCs from lung cancer patients and established a single-cell nanowell array for the molecular profiling of single CTCs [14]. They performed modular multigene panels, such as MET, telomerase reverse transcriptase (TERT), EGFR mutation, and EGFR wild-type, for the multigene profiling of individual CTCs from advanced-staged lung cancer patients to monitor therapy prediction and treatment. They detected positive samples (≥7 CTCs in 2 ml of blood) in 31 of 35 (88.6%) patients to identify CTCs using single-CTC MET and TERT expression profiling. Their nanoplatform demonstrated higher sensitivity for CTC identification than the CellSearch system, which only detected 53% of patients with advanced-staged lung cancer. They also performed EGFR mutation detection in single CTCs from patients who had a confirmed EGFR mutation status and observed heterogeneous levels of EGFR expression among CTCs. The MagSweeper system was also demonstrated to be capable of targeting metastasizing cells, thus offering a potential avenue to improve cancer therapy [71]. They measured high-purity gene expression in a single CTC without leukocyte contamination. The commonly expressed metastasis associated genes (e.g., S100A9, S100A4 and NPTN) and EMT genes (e.g., VIM, TGFß1, ZEB2, FOXC1 and CXCR4) of CTCs showed higher expression than tumor cell lines. Single-CTC profiling displayed a wide spectrum of gene differentiation that would be obscured when analyzing bulk CTCs. Therefore, they expected that single-cell transcriptional profiling of CTCs might facilitate the potential of 'liquid biopsy' for drug discovery. Other approaches such as laser microdissection and micropipette aspiration have also been demonstrated to be successful for single-CTC recovery. Hou et al. (Figure 6C) reported the next generation of NanoVelcro chips, which were prepared by depositing electrospun poly(lactic-co-glycolic acid) (PLGA) nanofibers onto a laser microdissection slide, thus allowing CTC analysis at single-cell resolution [120]. The targeted cells were first captured on anti-CD146-coated PLGA nanofibers, and a small piece of nanofiber was then cut using a 355-nm laser to collect single cells for subsequent molecular analysis. The genomic DNA (gDNA) extracted from individual CTCs was amplified using a commercial whole genome amplification kit to increase the amount of DNA, and the amplified gDNA was then subjected to Sanger sequencing to detect specific point mutations in melanoma patients (e.g., BRAFV600E mutation). Similarly, Zhao et al. reported a PLGA-modified NanoVelcro chip compatible with the same laser microdissection (LMD) technology for single-CTC isolation from prostate cancer patients [121]. The individually isolated CTCs from prostate cancer enabled whole exome sequencing (Exome-Seq) of pure CTCs, which may verify the role of CTCs as a tumor liquid biopsy and improve personalized medicine. The above LMD-induced cell release methods employed UV light that may cause cell DNA and RNA degradation. To release individual CTCs with high viability, Reátegui et al. (Figure 6D) demonstrated a dual-mode release mechanism that enabled the release of viable bulk or individual CTCs, making it successful for single-cell genotyping analysis (e.g., PIK3CA and epidermal growth factor receptor (EGFR) mutations) [122]. The device was fabricated via a layer-by-layer gelatin-coated nanostructure formation for effective CTC isolation (93% efficiency) and viable release (88% viability), which was achieved by temperature-responsive or mechano-sensitive release mechanisms. To confirm the clinical utility of the dual-mode nanoplatform, they performed single-CTC isolation and genotyping from two lung and three breast cancer patients who were confirmed to have EGFR and PIK3CA oncogenes, respectively. They demonstrated that all lung cancer patients were positive for the hotspot mutations of 2573T/G in the EGFR oncogene, and all individual CTCs captured from breast cancer patients were identified with 3140A/G mutations in the PIK3CA oncogene.
Potential clinical application of CTCs
CTCs may play a key role in cancer metastasis and be responsible for the lethality of most cancers. As a real-time liquid biopsy, CTCs potentially hold clinical utility in their ability to aid in treatment decisions [123-125]. Given that blood samples can be obtained conveniently, this liquid biopsy will be beneficial for monitoring drug resistance and clinical response. However, a CTC-based point-of-care test remains difficult to achieve due to the inadequate selectivity and sensitivity for CTC isolation. Nanotechnologies are able to solve the problems of inadequate efficiency and purity of CTCs owing to the unique physical and chemical properties of nanomaterials, enabling the further understanding and processing of CTCs, including enumeration and molecular profiling. The number of CTCs reflects tumor aggressiveness and the ultimate cancer patient outcome, as the detection of five or more CTCs in 7.5 ml blood samples is associated with adverse clinical outcomes [126]. Uncovering CTC phenotypes provides the potential for understanding the biology of metastasis and drug resistance [127, 128]. Improved diagnostic accuracy is important for optimal clinical decision making in order to match the patient with an appropriate treatment.
CTC analysis includes enumeration, identification, and characterization. Presently, the widely accepted identification and enumeration of CTCs based on immunofluorescence staining, using fluorescent antibodies to target biological markers, involves the expression of epithelial markers such as cytokeratins and EpCAM without the presence of the leukocyte marker CD45 [129]. In addition, CTCs can also be identified and enumerated based on the expression of mesenchymal markers (e.g., vimentin) [130], stem cell markers (e.g., CD133) [131], or a shift in the expression of epithelial markers (e.g., E-cadherin) to mesenchymal markers (e.g., N-cadherin)[132]. Various enumeration and detection methods have been used to analyze cancer patient samples and evaluate the clinical significance of CTCs, such as the prognosis of cancer patients and treatment response monitoring [133]. For example, Nagrath and co-workers assessed the clinical value of CTC enumeration for real-time monitoring of responses to therapy [93]. They demonstrated that CTC quantity correlates reasonably well with clinical response after a small cohort of clinical samples were analyzed, including patients with pancreatic, colorectal, esophageal, and non-small cell lung cancer. Liu et al. carried out a study with 74 breast cancer patients confirming the value of CTC counts as an aid to standard methods for monitoring disease progression [134]. Serial CTC levels were obtained in metastatic breast cancer patients undergoing endocrine or chemotherapy therapy, and >5 CTCs per 7.5 ml of blood was found to predict poorer progression-free survival. However, CTC enumeration alone may not provide more detailed information for monitoring disease development and discovering new biomarkers for CTC detection. Moreover, CTC enumeration as a test of disease status is completely based on sensitive CTC detection, making it unable to monitor disease processes.
Downstream genetic analyses of CTCs have long been explored to provide deeper insights into CTCs [135, 136]. Technologies such as fluorescent in situ hybridization (FISH) [137], comparative genomic hybridization (CGH) [138], DNA whole genome amplification (WGA) for gene sequencing [120], and DNA transcriptional analysis (e.g., quantitative real-time polymerase chain reaction, qRT-PCR) have been employed in research to analyze the molecular properties of CTCs [139]. FISH is a fluorescence-based technology demonstrated as an effective method for CTC genotyping by detecting specific DNA sequences on chromosomes. FISH has been employed to characterize the invasive potential of CTCs by evaluating the overexpression of tumor markers on enriched cells, such as human epidermal growth factor receptor 2 (HER2), androgen receptor (AR) and EGFR [140, 141]. Among these biomarkers, HER2 is widely used as a clinical molecular therapeutic target for 30% of gastric and breast cancer patients [142, 143]. The status of HER2 in CTCs is beneficial for the determination of an appropriate treatment for cancer patients. For example, Meng et al. evaluated the HER2 gene expression of CTCs isolated from 24 breast cancer patients, and showed that the primary tumor was HER2-negative and 9 patients had HER2 gene amplification in their CTCs [144]. They also demonstrated that CTCs could represent a stable real-time liquid biopsy to detect genetic changes as cancer progresses and could reflect the status of the recurrent tumors. An alternative method frequently used in analyzing oncogene mutations in CTCs is CGH, which is utilized to assess copy number variations. Furthermore, the combination of single-cell isolation techniques with WGA methods has allowed the analysis of multiple mutations in individual cells. These methods identify copy number changes (e.g., losses and gains) in the genome of a single CTC at high resolution, and these high-level amplifications might become valuable future candidates for treatment. Similarly, PCR-based methods can be applied to qualitatively and quantitatively determine the expression level of specific target sequences and amplify tumor-specific deformities present in the mRNA or DNA. AmpliGrid (Beckman Coulter Genomics) is a PCR-based chip for individual cell deposition and PCR analysis [145], which allows the genetic determination of cell heterogeneity, microRNA levels, genetic fingerprint, or epigenetic features. The capability of RNA to reveal the origin/mutations of tissues that are associated with tumor development makes it an important resource in CTC survey [146]. Thus, RNA separation and analysis are areas of interest that can be improved by the application of nanotechnologies. For example, a microvortex-generating herringbone chip was developed to detect CTCs from prostate cancer patients and investigate the molecular characterization of CTCs [147]. The enriched cells were lysed on-chip to isolate a sufficient quantity of RNA for genetic analysis. Ivanov et al. developed a multiplexed sensor chip to analyze and classify prostate cancer cells [148]. A nanostructured coating of palladium was deposited on the surface of the electrodes to increase the performance of the probes. Following extraction of RNA from isolated CTCs, it was able to identify the cells that originated from prostate cancer using electrodes coated with peptide nucleic acid probes against the TMPRSS/ERG Type III gene fusion or prostate-specific antigen (PSA) gene.
The analysis of CTCs from molecular and genetic perspectives provides more accurate and detailed descriptions regarding cancer metastasis compared with CTC enumeration. CTCs may develop mutations during the cancer metastasis process and drug treatment. Cancer cells undergoing EMT are considered more malignant and drug resistant [149]. Researchers have worked on the characterization of these EMT-induced EpCAM-negative CTCs for further investigation of cancer metastasis and therapeutic resistance. To investigate the role of EMT in cancer patients, Yu et al. used an HB-chip to capture CTCs and analyzed EMT in CTCs from breast cancer patients [150]. They established an RNA-in situ hybridization assay (RNA-ISH) to examine the dynamic changes in epithelial and mesenchymal composition. They found that mesenchymal CTCs occurred as both clusters and single cells, and transforming growth factor (TGF)-β activation and aberrant expression of Forkhead box protein C1 (FOXC1) may contribute to EMT.
Enumeration and molecular analyses of CTCs pave the way for improved understanding of cancer metastasis and to provide potential treatments for cancer. To achieve more accurate information, nanotechnologies involved in CTC capture and detection must be optimized. An ideal CTC capture platform should be able to isolate rare and heterogeneous CTCs to prevent potential false negative signals, which is a technical challenge. Nanomaterials and microchip-based technologies have the potential to accelerate the bio-analysis of CTCs.
Conclusions and perspectives
In this review, we categorize and discuss types of nanotechniques used to isolate and analyze CTCs as well as their clinical applications. To date, technologies for CTC isolation suffer from one or more limitations, such as low sensitivity, low purity, low throughput, inability to release fixed cells, and dependence on expensive instruments for enrichment or subsequent characterization. Because of their high area-to-volume ratio and similar scales to cellular pseudopodia, nanomaterials (e.g., magnetic nanoparticles, nanowires, nanopillars, carbon nanotubes and GO) have been applied to solve the challenges of the low capture efficiency and insufficient purity of CTCs. Nanomaterials are ideal platforms for multiplexed targeting because they can be modified with different targeting ligands to capture and analyze CTC subpopulations. Nanomaterials can also be embedded into microfluidic devices to facilitate CTC isolation. Due to the advantages of parameter manipulation and accurate flow control and the benefit of synergistic effects when nanomaterials are incorporated, microfluidics are versatile devices for achieving high specificity, yield and purity in CTC isolation.
CTC isolation technologies are primarily based on the characteristics of CTCs that distinguish them from the surrounding normal blood cells, including physical and biological properties. Physical CTC isolation platforms are generally simpler to produce but typically exhibit low CTC purity. Biological CTC isolation platforms are capable of higher CTC selection purity, making these strategies suitable for molecular analyses, such as FISH, CGH, WGA and mRNA expression profiling. However, biological CTC assays are also limited by requiring robust capture agent immobilization chemistries and release strategies. In addition, the present assays based on epithelial antigens (e.g., cytokeratins or EpCAM) may miss the most invasive CTC subpopulations, such as CTCs that lose epithelial antigen expression when cancer cells undergo EMT. In fact, because EMT is a necessary part of metastasis, the subpopulation of CTCs that have decreased expression of epithelial markers are actually the most important prognostic factors for disease. Thus, there is an urgent need to develop a better strategy for isolating and identifying EMT-like subsets of tumor cells.
The detection and characterization of CTCs will provide new insights into the mechanism of cancer metastasis and into the clinical management of cancers. However, because of the low quantity of CTCs detected by the currently available methods, their value as a “liquid biopsy” is limited, particularly in early-stage cancer patients. Specificity and sensitivity remain the key issues that future technologies need to address. For CTC enumeration, the key parameter is yield, which must be as high as possible to accurately define the concentration of these cells in the blood of patients. Invasion may occur early in tumor development, thereby raising the issue of the application of highly specific and sensitive methods for the detection of CTCs in cancer patients. For the molecular characterization of CTCs, such as using genome sequencing and gene expression analysis, yield is not the only metric of significance. Instead, sample purity is extremely important due to the need for a high-purity template with minimal contamination. Molecular profiling of CTCs using clinically relevant genetic markers (e.g., HER2, KRAS, and EGFR) will require technologies that can generate high-purity CTC isolates [142, 151-153], whereas single-cell assays can relax the need for high purity to accommodate follow-up molecular assays to adapt treatment regimens.
Studies regarding the predictive and prognostic value of CTCs are still under active investigation. Thus far, the CellSearch system is the only FDA-approved cell capture platform, and it only detects a few types of cancer. More technologies should be translated for clinical use to achieve clinical validity and utility. In addition to high efficiency, high throughput and high purity, improvements in cell integrity and post-isolation cell cultures are also urgently needed for future devices focused on CTC isolation. In addition, a cascaded isolation system may allow for cancer research and drug screening. CTCs could originate from the primary tumor or from any metastatic sites, which might result in heterogeneity of cells. To determine the relationships between the heterogeneous populations of CTCs, a tumor-specific CTC platform is an urgent need in place of one technology for CTC detection in all types of cancer. In addition, due to the intrinsic heterogeneity of tumors and the cells undergoing EMT, the use of a single capture agent assay cannot provide sufficient clinical sensitivity. Aptamers, for example, could offer the best specificity for a variety of target CTCs and can be further explored as effective new markers for exploring the invasive potential of CTCs and guiding anticancer treatments. The integration of CTC analyses with emerging diagnostics that rely on circulating cell-free nucleic acids (microRNA, mRNA and DNA) and exosomes will also be explored to provide complementary information for cancer screening and prognosis and the monitoring of anticancer therapy efficacy [154-158].
Acknowledgements
Hubei Province's Outstanding Medical Academic Leader Program, and the Young One Thousand Talent Program of China.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Eheman C, Henley SJ, Ballard-Barbash R, Jacobs EJ, Schymura MJ, Noone AM. et al. Annual Report to the Nation on the status of cancer, 1975-2008, featuring cancers associated with excess weight and lack of sufficient physical activity. Cancer. 2012;118:2338-66
2. Bhana S, Wang Y, Huang X. Nanotechnology for enrichment and detection of circulating tumor cells. Nanomedicine. 2015;10:1973
3. Aceto N, Bardia A, Miyamoto David T, Donaldson Maria C, Wittner Ben S, Spencer Joel A. et al. Circulating Tumor Cell Clusters Are Oligoclonal Precursors of Breast Cancer Metastasis. Cell. 2014;158:1110-22
4. Ameri K, Luong R, Zhang H, Powell AA, Montgomery KD, Espinosa I. et al. Circulating tumour cells demonstrate an altered response to hypoxia and an aggressive phenotype. Br J Cancer. 2010;102:561-9
5. Chaffer CL, Weinberg RA. A Perspective on Cancer Cell Metastasis. Science. 2011;331:1559-64
6. Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med. 2006;12:895-904
7. Marx V. Tracking metastasis and tricking cancer. Nature. 2013;494:133-8
8. Jin XR, Zhu LY, Qian K, Feng YG, Zhou JH, Wang RW. et al. Circulating tumor cells in early stage lung adenocarcinoma: a case series report and literature review. Oncotarget. 2017;8:23130-41
9. Plaks V, Koopman CD, Werb Z. Circulating Tumor Cells. Science. 2013;341:1186-8
10. Banys-Paluchowski M, Krawczyk N, Fehm T. Potential Role of Circulating Tumor Cell Detection and Monitoring in Breast Cancer: A Review of Current Evidence. Front Oncol. 2016;6:255
11. Pantel K, Alix-Panabières C. Real-time Liquid Biopsy in Cancer Patients: Fact or Fiction? Cancer Research. 2013;73:6384-8
12. Kling J. Beyond counting tumor cells. Nat Biotech. 2012;30:578-80
13. Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: approaches to isolation and characterization. The Journal of Cell Biology. 2011;192:373-82
14. Park SM, Wong DJ, Ooi CC, Kurtz DM, Vermesh O, Aalipour A. et al. Molecular profiling of single circulating tumor cells from lung cancer patients. Proc Natl Acad Sci U S A. 2016;113:E8379-E86
15. Laget S, Broncy L, Hormigos K, Dhingra DM, BenMohamed F, Capiod T. et al. Technical Insights into Highly Sensitive Isolation and Molecular Characterization of Fixed and Live Circulating Tumor Cells for Early Detection of Tumor Invasion. PLoS One. 2017;12:e0169427
16. Souza ESV, Chinen LT, Abdallah EA, Damascena A, Paludo J, Chojniak R. et al. Early detection of poor outcome in patients with metastatic colorectal cancer: tumor kinetics evaluated by circulating tumor cells. Onco Targets Ther. 2016;9:7503-13
17. Wang H, Stoecklein NH, Lin PP, Gires O. Circulating and disseminated tumor cells: diagnostic tools and therapeutic targets in motion. Oncotarget. 2017;8:1884-912
18. Hong B, Zu Y. Detecting circulating tumor cells: current challenges and new trends. Theranostics. 2013;3:377-94
19. Toom EEVD, Verdone JE, Gorin MA, Pienta KJ. Technical challenges in the isolation and analysis of circulating tumor cells. Oncotarget. 2016;7:62754-66
20. Pantel K, Alix-Panabières C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends in molecular medicine. 2010;16:398-406
21. Bulfoni M, Turetta M, Ben FD, Loreto CD, Beltrami AP, Cesselli D. Dissecting the Heterogeneity of Circulating Tumor Cells in Metastatic Breast Cancer: Going Far Beyond the Needle in the Haystack. Int J Mol Sci. 2016;17:1775
22. Kelley SO, Mirkin CA, Walt DR, Ismagilov RF, Toner M, Sargent EH. Advancing the speed, sensitivity and accuracy of biomolecular detection using multi-length-scale engineering. Nat Nano. 2014;9:969-80
23. Liu Y, Ling Y, Qi Q, Lan F, Zhu M, Zhang Y. et al. Prognostic value of circulating tumor cells in advanced gastric cancer patients receiving chemotherapy. Mol Clin Oncol. 2017;6:235-42
24. Schroeder A, Heller DA, Winslow MM, Dahlman JE, Pratt GW, Langer R. et al. Treating metastatic cancer with nanotechnology. Nat Rev Cancer. 2012;12:39-50
25. Ming Y, Li Y, Xing H, Luo M, Li Z, Chen J. et al. Circulating Tumor Cells: From Theory to Nanotechnology-Based Detection. Front Pharmacol. 2017;8:35
26. Zhang JJ, Khademhosseini A. Emerging micro- and nanotechnologies in cancer diagnosis and therapy. Biomedical Microdevices. 2013;15:579-81
27. Hughes AD, King MR. Nanobiotechnology for the capture and manipulation of circulating tumor cells. Wiley Interdisciplinary Reviews: Nanomedicine and Nanobiotechnology. 2012;4:291-309
28. Chen W, Allen SG, Reka AK, Qian W, Han S, Zhao J. et al. Nanoroughened adhesion-based capture of circulating tumor cells with heterogeneous expression and metastatic characteristics. BMC Cancer. 2016;16:614
29. Chen W, Weng S, Zhang F, Allen S, Li X, Bao L. et al. Nanoroughened Surfaces for Efficient Capture of Circulating Tumor Cells without Using Capture Antibodies. Acs Nano. 2013;7:566-75
30. Wilson R. The use of gold nanoparticles in diagnostics and detection. Chemical Society Reviews. 2008;37:2028-45
31. Yoon HJ, Kim TH, Zhang Z, Azizi E, Pham TM, Paoletti C. et al. Sensitive capture of circulating tumour cells by functionalized graphene oxide nanosheets. Nat Nano. 2013;8:735-41
32. Panchapakesan B, Cesarone G, Liu S, Teker K, Wickstrom E. Single-wall carbon nanotubes with adsorbed antibodies detect live breast cancer cells. Nanobiotechnology. 2005;1:353-60
33. Min H, Jo SM, Kim HS. Efficient capture and simple quantification of circulating tumor cells using quantum dots and magnetic beads. Small. 2015;11:2536-42
34. Fang S, Wang C, Xiang J, Cheng L, Song X, Xu L. et al. Aptamer-conjugated upconversion nanoprobes assisted by magnetic separation for effective isolation and sensitive detection of circulating tumor cells. Nano Research. 2014;7:1327-36
35. Pan Y, Du X, Zhao F, Xu B. Magnetic nanoparticles for the manipulation of proteins and cells. Chemical Society Reviews. 2012;41:2912-42
36. Xiong B, Ren K, Shu Y, Chen Y, Shen B, Wu H. Recent Developments in Microfluidics for Cell Studies. Advanced Materials. 2014;26:5525-32
37. Chen Y, Li P, Huang P-H, Xie Y, Mai JD, Wang L. et al. Rare cell isolation and analysis in microfluidics. Lab on a Chip. 2014;14:626-45
38. Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442:368-73
39. Mu X, Zheng W, Sun J, Zhang W, Jiang X. Microfluidics for Manipulating Cells. Small. 2013;9:9-21
40. Yu ZTF, Aw Yong KM, Fu J. Microfluidic Blood Cell Sorting: Now and Beyond. Small. 2014;10:1687-703
41. Li P, Stratton ZS, Dao M, Ritz J, Huang TJ. Probing circulating tumor cells in microfluidics. Lab on A Chip. 2013;13:602-9
42. Lin M, Chen J-F, Lu Y-T, Zhang Y, Song J, Hou S. et al. Nanostructure Embedded Microchips for Detection, Isolation, and Characterization of Circulating Tumor Cells. Accounts of Chemical Research. 2014;47:2941-50
43. Yu L, Ng SR, Xu Y, Dong H, Wang YJ, Li CM. Advances of lab-on-a-chip in isolation, detection and post-processing of circulating tumour cells. Lab on a Chip. 2013
44. Kim MS, Sim TS, Kim YJ, Kim SS, Jeong H, Park J-M. et al. SSA-MOA: a novel CTC isolation platform using selective size amplification (SSA) and a multi-obstacle architecture (MOA) filter. Lab on a Chip. 2012;12:2874-80
45. Koh Y, Kenmotsu H, Hosokawa M, Yoshino T, Naito T, Watanabe R. et al. 260 Deformability-based Isolation of Circulating Tumor Cells in Lung Cancer Patients with Microcavity Array System. Eur J Cancer. 2012;48:80 -
46. Gertler R, Rosenberg R, Fuehrer K, Dahm M, Nekarda H, Siewert JR. Detection of circulating tumor cells in blood using an optimized density gradient centrifugation. Recent Results Cancer Res. 2003;162:149
47. Shim S, Stemkehale K, Tsimberidou AM, Noshari J, Anderson TE, Gascoyne PR. Antibody-independent isolation of circulating tumor cells by continuous-flow dielectrophoresis. Biomicrofluidics. 2013;7:S38-S40
48. Chem A. Size-Selective Microcavity Array for Rapid and Efficient Detection of Circulating Tumor Cells. Analytical Chemistry. 2010;82:6629
49. Lin HK, Zheng S, Williams AJ, Balic M, Groshen S, Scher HI. et al. Portable Filter-Based Microdevice for Detection and Characterization of Circulating Tumor Cells. Clinical Cancer Research. 2010;16:5011-8
50. Autebert J, Coudert B, Bidard FC, Pierga JY, Descroix S, Malaquin L. et al. Microfluidic: An innovative tool for efficient cell sorting. Methods. 2012;57:297-307
51. Li Y, Dalton C, Crabtree HJ, Nilsson G, Kaler KVIS. Continuous dielectrophoretic cell separation microfluidic device. Lab on a Chip. 2007;7:239-48
52. Thege FI, Lannin TB, Saha TN, Tsai S, Kochman ML, Hollingsworth MA. et al. Microfluidic immunocapture of circulating pancreatic cells using parallel EpCAM and MUC1 capture: characterization, optimization and downstream analysis. Lab on A Chip. 2014;14:1775-84
53. Kaiser J. Cancer's Circulation Problem. Science. 2010;327:1072-4
54. Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC. et al. Circulating Tumor Cells, Disease Progression, and Survival in Metastatic Breast Cancer. New England Journal of Medicine. 2004;351:781-91
55. Pimienta M, Edderkaoui M, Wang R, Pandol S. The Potential for Circulating Tumor Cells in Pancreatic Cancer Management. Front Physiol. 2017;8:381
56. Alix-Panabieres C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer. 2014;14:623-31
57. Kölbl AC, Jeschke U, Andergassen U. The Significance of Epithelial-to-Mesenchymal Transition for Circulating Tumor Cells. Int J Mol Sci. 2016;17:1308
58. Sharma TK, Bruno JG, Dhiman A. ABCs of DNA aptamer and related assay development. Biotechnology Advances. 2017;35:275-301
59. Wang S, Zhang C, Wang G, Cheng B, Wang Y, Chen F. et al. Aptamer-Mediated Transparent-Biocompatible Nanostructured Surfaces for Hepotocellular Circulating Tumor Cells Enrichment. Theranostics. 2016;6:1877-86
60. Hassan EM, Willmore WG, DeRosa MC. Aptamers: Promising Tools for the Detection of Circulating Tumor Cells. Nucleic acid therapeutics. 2016;26:335-47
61. Pantel K, Alix-Panabieres C. Technologies for detection of circulating tumor cells: Facts and Vision. Lab on a Chip. 2013
62. Yoon HJ, Kozminsky M, Nagrath S. Emerging Role of Nanomaterials in Circulating Tumor Cell Isolation and Analysis. ACS Nano. 2014;8:1995-2017
63. Liu X, Wang S. Three-dimensional nano-biointerface as a new platform for guiding cell fate. Chemical Society Reviews. 2014;43:2385-401
64. Sekine J, Luo S-C, Wang S, Zhu B, Tseng H-R, Yu H-h. Functionalized Conducting Polymer Nanodots for Enhanced Cell Capturing: The Synergistic Effect of Capture Agents and Nanostructures. Advanced Materials. 2011;23:4788-92
65. Dong-Joo K, Jin-Kyeong S, Geehee L, Gil-Sung K, Sang-Kwon L. Cell adhesion and migration on nanopatterned substrates and their effects on cell-capture yield. Nanotechnology. 2012;23:395102
66. Wang S, Wang H, Jiao J, Chen K-J, Owens GE, Kamei K-i. et al. Three-Dimensional Nanostructured Substrates toward Efficient Capture of Circulating Tumor Cells. Angewandte Chemie. 2009;121:9132-5
67. Zhang N, Deng Y, Tai Q, Cheng B, Zhao L, Shen Q. et al. Electrospun TiO2 nanofiber-based cell capture assay for detecting circulating tumor cells from colorectal and gastric cancer patients. Adv Mater. 2012;24:2756-60
68. Zheng F, Cheng Y, Wang J, Lu J, Zhang B, Zhao Y. et al. Aptamer-Functionalized Barcode Particles for the Capture and Detection of Multiple Types of Circulating Tumor Cells. Advanced Materials. 2014;26:7333-8
69. Pankhurst QA, Connolly J, Jones SK, Dobson J. Applications of magnetic nanoparticles in biomedicine. Journal of Physics D: Applied Physics. 2003;36:R167
70. Miltenyi S, Müller W, Weichel W, Radbruch A. High gradient magnetic separation with MACS. Cytometry. 1990;11:231-8
71. Powell AA, Talasaz AH, Zhang H, Coram MA, Reddy A, Deng G. et al. Single cell profiling of circulating tumor cells: transcriptional heterogeneity and diversity from breast cancer cell lines. PLoS One. 2012;7:e33788
72. Huang C, Yang G, Ha Q, Meng J, Wang S. Multifunctional “Smart” Particles Engineered from Live Immunocytes: Toward Capture and Release of Cancer Cells. Advanced Materials. 2015;27:310-3
73. Hultgren A, Tanase M, Chen CS, Meyer GJ, Reich DH. Cell manipulation using magnetic nanowires. Journal of Applied Physics. 2003;93:7554-6
74. Hultgren A, Tanase M, Chen CS, Reich DH. High-Yield Cell Separations Using Magnetic Nanowires. IEEE Transactions on Magnetics. 2004;40:2988-90
75. Hultgren A, Tanase M, Felton EJ, Bhadriraju K, Salem AK, Chen CS. et al. Optimization of Yield in Magnetic Cell Separations Using Nickel Nanowires of Different Lengths. Biotechnology Progress. 2008;21:509-15
76. Hong W, Lee S, Chang HJ, Lee ES, Cho Y. Multifunctional magnetic nanowires: A novel breakthrough for ultrasensitive detection and isolation of rare cancer cells from non-metastatic early breast cancer patients using small volumes of blood. Biomaterials. 2016;106:78-86
77. Orndorff RL, Rosenthal SJ. Neurotoxin Quantum Dot Conjugates Detect Endogenous Targets Expressed in Live Cancer Cells. Nano Letters. 2009;9:2589-99
78. Orte A, Alvarezpez JM, Ruedasrama MJ. Fluorescence Lifetime Imaging Microscopy for the Detection of Intracellular pH with Quantum Dot Nanosensors. Acs Nano. 2013;7:6387-95
79. Lee H, Kim C, Lee D, Park JH, Searson PC, Lee KH. Optical coding of fusion genes using multicolor quantum dots for prostate cancer diagnosis. Int J Nanomedicine. 2017;12:4397-407
80. Lee J, Kang HJ, Jang H, Lee YJ, Lee YS, Ali BA. et al. Simultaneous imaging of two different cancer biomarkers using aptamer-conjugated quantum dots. Sensors (Basel). 2015;15:8595-604
81. Xie M, Lu N-N, Cheng S-B, Wang X-Y, Wang M, Guo S. et al. Engineered Decomposable Multifunctional Nanobioprobes for Capture and Release of Rare Cancer Cells. Analytical Chemistry. 2014;86:4618-26
82. Huang X, O'Connor R, Kwizera EA. Gold Nanoparticle Based Platforms for Circulating Cancer Marker Detection. Nanotheranostics. 2017;1:80
83. Zhou W, Gao X, Liu D, Chen X. Gold Nanoparticles for In Vitro Diagnostics. Chem Rev. 2015;115:10575-636
84. Maltez-da Costa M, de la Escosura-Muniz A, Nogues C, Barrios L, Ibanez E, Merkoci A. Simple monitoring of cancer cells using nanoparticles. Nano Lett. 2012;12:4164-71
85. Zhu Y, Chandra P, Shim Y-B. Ultrasensitive and Selective Electrochemical Diagnosis of Breast Cancer Based on a Hydrazine-Au Nanoparticle-Aptamer Bioconjugate. Analytical Chemistry. 2013;85:1058-64
86. Yi Z, Li XY, Gao Q, Tang LJ, Chu X. Aptamer-aided target capturing with biocatalytic metal deposition: an electrochemical platform for sensitive detection of cancer cells. Analyst. 2013;138:2032-7
87. Zhang Y, Chen B, He M, Yang B, Zhang J, Hu B. Immunomagnetic Separation Combined with Inductively Coupled Plasma Mass Spectrometry for the Detection of Tumor Cells Using Gold Nanoparticle Labeling. Analytical Chemistry. 2014;86:8082-9
88. Liu Y, Zhu F, Dan W, Fu Y, Liu S. Construction of carbon nanotube based nanoarchitectures for selective impedimetric detection of cancer cells in whole blood. Analyst. 2014;139:5086-92
89. Jackson JM, Witek MA, Kamande JW, Soper SA. Materials and microfluidics: enabling the efficient isolation and analysis of circulating tumour cells. Chemical Society Reviews. 2017;46:4245-80
90. Qian W, Zhang Y, Chen W. Capturing Cancer: Emerging Microfluidic Technologies for the Capture and Characterization of Circulating Tumor Cells. Small. 2015;11:3850-72
91. Wang S, Liu K, Liu J, Yu ZTF, Xu X, Zhao L. et al. Highly Efficient Capture of Circulating Tumor Cells by Using Nanostructured Silicon Substrates with Integrated Chaotic Micromixers. Angewandte Chemie International Edition. 2011;50:3084-8
92. Yu X, He R, Li S, Cai B, Zhao L, Liao L. et al. Magneto-Controllable Capture and Release of Cancer Cells by Using a Micropillar Device Decorated with Graphite Oxide-Coated Magnetic Nanoparticles. Small. 2013;9:3895-901
93. Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L. et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235-9
94. Sheng W, Chen T, Tan W, Fan ZH. Multivalent DNA Nanospheres for Enhanced Capture of Cancer Cells in Microfluidic Devices. Acs Nano. 2013;7:7067-76
95. Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA. et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18392-7
96. Wang S, Sohrabi S, Xu J, Yang J, Liu Y. Geometry design of herringbone structures for cancer cell capture in a microfluidic device. Microfluidics and Nanofluidics. 2016:20
97. Shi W, Wang S, Maarouf A, Uhl CG, He R, Yunus D. et al. Magnetic particles assisted capture and release of rare circulating tumor cells using wavy-herringbone structured microfluidic devices. Lab Chip. 2017
98. Chen P, Huang YY, Hoshino K, Zhang X. Multiscale immunomagnetic enrichment of circulating tumor cells: from tubes to microchips. Lab on A Chip. 2013;14:446-58
99. Earhart CM, Hughes CE, Gaster RS, Ooi CC, Wilson RJ, Zhou LY. et al. Isolation and mutational analysis of circulating tumor cells from lung cancer patients with magnetic sifters and biochips. Lab on A Chip. 2013;14:78-88
100. Hoshino K, Huang YY, Lane N, Huebschman M, Uhr JW, Frenkel EP. et al. Microchip-based immunomagnetic detection of circulating tumor cells. Lab Chip. 2011;11:3449-57
101. Chen CL, Chen KC, Pan YC, Lee TP, Hsiung LC, Lin CM. et al. Separation and detection of rare cells in a microfluidic disk via negative selection. Lab Chip. 2011;11:474-83
102. Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E. et al. Inertial Focusing for Tumor Antigen-Dependent and -Independent Sorting of Rare Circulating Tumor Cells. Science Translational Medicine. 2013;5:179ra47
103. Mohamadi RM, Besant JD, Mepham A, Green B, Mahmoudian L, Gibbs T. et al. Nanoparticle-Mediated Binning and Profiling of Heterogeneous Circulating Tumor Cell Subpopulations. Angewandte Chemie International Edition. 2015;54:139-43
104. Poudineh M, Aldridge PM, Ahmed S, Green BJ, Kermanshah L, Nguyen V. et al. Tracking the dynamics of circulating tumour cell phenotypes using nanoparticle-mediated magnetic ranking. Nat Nanotechnol. 2017;12:274-81
105. Sosa MS, Bragado P, Aguirre-Ghiso JA. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat Rev Cancer. 2014;14:611-22
106. Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat Rev Cancer. 2008;8:329-40
107. Hou S, Zhao H, Zhao L, Shen Q, Wei KS, Suh DY. et al. Capture and stimulated release of circulating tumor cells on polymer-grafted silicon nanostructures. Advanced Materials. 2013;25:1547-51
108. Huang Q, Chen B, He R, He Z, Cai B, Xu J. et al. Capture and release of cancer cells based on sacrificeable transparent MnO2 nanospheres thin film. Adv Healthc Mater. 2014;3:1420-5
109. Huang Q, Cai B, Chen B, Rao L, He Z, He R. et al. Efficient Purification and Release of Circulating Tumor Cells by Synergistic Effect of Biomarker and SiO2 @Gel-Microbead-Based Size Difference Amplification. Adv Healthc Mater. 2016;5:1554-9
110. Shen Q, Xu L, Zhao L, Wu D, Fan Y, Zhou Y. et al. Specific capture and release of circulating tumor cells using aptamer-modified nanosubstrates. Advanced Materials. 2013;25:2368-73
111. Lee HJ, Cho HY, Oh JH, Namkoong K, Lee JG, Park JM. et al. Simultaneous capture and in situ analysis of circulating tumor cells using multiple hybrid nanoparticles. Biosens Bioelectron. 2013;47:508-14
112. Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC. et al. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345:216-20
113. Chen XX, Bai F. Single-cell analyses of circulating tumor cells. Cancer biology & medicine. 2015;12:184-92
114. Wang D, Bodovitz S. Single cell analysis: the new frontier in 'omics'. Trends Biotechnol. 2010;28:281-90
115. Miyamoto DT, Ting DT, Toner M, Maheswaran S, Haber DA. Single-Cell Analysis of Circulating Tumor Cells as a Window into Tumor Heterogeneity. Cold Spring Harb Symp Quant Biol. 2016;81:269-74
116. Jordan NV, Bardia A, Wittner BS, Benes C, Ligorio M, Zheng Y. et al. HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature. 2016;537:102-6
117. Bingham C, Fernandez SV, Fittipaldi P, Dempsey PW, Ruth KJ, Cristofanilli M. et al. Mutational studies on single circulating tumor cells isolated from the blood of inflammatory breast cancer patients. Breast Cancer Res Treat. 2017;163:219-30
118. Zheng Y, Nguyen J, Wei Y, Sun Y. Recent advances in microfluidic techniques for single-cell biophysical characterization. Lab on A Chip. 2013;13:2464-83
119. Yeo T, Tan SJ, Lim CL, Lau DP, Chua YW, Krisna SS. et al. Microfluidic enrichment for the single cell analysis of circulating tumor cells. Scientific reports. 2016;6:22076
120. Hou S, Zhao L, Shen Q, Yu J, Ng C, Kong X. et al. Polymer Nanofiber-Embedded Microchips for Detection, Isolation, and Molecular Analysis of Single Circulating Melanoma Cells. Angewandte Chemie International Edition. 2013;52:3379-83
121. Zhao L, Lu YT, Li F, Wu K, Hou S, Yu J. et al. High-Purity Prostate Circulating Tumor Cell Isolation by a Polymer Nanofiber-Embedded Microchip for Whole Exome Sequencing. Advanced Materials. 2013;25:2897-902
122. Reátegui E, Aceto N, Lim EJ, Sullivan JP, Jensen AE, Zeinali M. et al. Tunable Nanostructured Coating for the Capture and Selective Release of Viable Circulating Tumor Cells. Advanced Materials. 2015;27:1593-9
123. Arya SK, Lim B, Rahman ARA. Enrichment, detection and clinical significance of circulating tumor cells. Lab on a Chip. 2013;13:1995-2027
124. Z B, A J, M P, M Z, K K, V B. Circulating tumor cells: what we know, what do we want to know about them and are they ready to be used in clinics? American journal of translational research. 2017;9:2807
125. Wu ZX, Liu Z, Jiang HL, Pan HM, Han WD. Circulating tumor cells predict survival benefit from chemotherapy in patients with lung cancer. Oncotarget. 2016;7:67586-96
126. Masuda T, Hayashi N, Iguchi T, Ito S, Eguchi H, Mimori K. Clinical and biological significance of circulating tumor cells in cancer. Molecular oncology. 2016;10:408-17
127. Luo X, Mitra D, Sullivan RJ, Wittner BS, Kimura AM, Pan S. et al. Isolation and molecular characterization of circulating melanoma cells. Cell Rep. 2014;7:645-53
128. Valastyan S, Weinberg Robert A. Tumor Metastasis: Molecular Insights and Evolving Paradigms. Cell. 2011;147:275-92
129. Xu Y, Qin T, Li J, Wang X, Gao C, Xu C. et al. Detection of Circulating Tumor Cells Using Negative Enrichment Immunofluorescence and an In Situ Hybridization System in Pancreatic Cancer. Int J Mol Sci. 2017;18:622
130. Yokobori T, Iinuma H, Shimamura T, Imoto S, Sugimachi K, Ishii H. et al. Plastin3 Is a Novel Marker for Circulating Tumor Cells Undergoing the Epithelial-Mesenchymal Transition and Is Associated with Colorectal Cancer Prognosis. Cancer Research. 2013;73:2059-69
131. Yang B, Wang Y, Yang C, Ouyang W, Zhou F, Zhou Y. et al. The Ultrastructural Difference between CD133-positive U251 Glioma Stem Cells and Normal U251 Glioma Cells. Ultrastructural Pathology. 2012;36:404-8
132. Rhim Andrew D, Mirek Emily T, Aiello Nicole M, Maitra A, Bailey Jennifer M, McAllister F. et al. EMT and Dissemination Precede Pancreatic Tumor Formation. Cell. 2012;148:349-61
133. Zhou J, Ma X, Bi F, Liu M. Clinical significance of circulating tumor cells in gastric cancer patients. Oncotarget. 2017;8:25713-20
134. Liu MC, Shields PG, Warren RD, Cohen P, Wilkinson M, Ottaviano YL. et al. Circulating tumor cells: a useful predictor of treatment efficacy in metastatic breast cancer. J Clin Oncol. 2009;27:5153-9
135. Lack J, Gillard M, Cam M, Paner GP, VanderWeele DJ. Circulating tumor cells capture disease evolution in advanced prostate cancer. J Transl Med. 2017;15:44
136. Niravath P, Cakar B, Ellis M. The role of genetic testing in the selection of therapy for breast cancer: A review. JAMA oncology. 2017;3:262-8
137. Wei W, Zhang Z, Xian HG, Zhen S, Yan J, Lu H. et al. Clinical significance of detecting circulating tumor cells in colorectal cancer using subtraction enrichment and immunostaining-fluorescence in situ hybridization (SE-iFISH). Oncotarget. 2017;8:21639-49
138. Inazawa J, Inoue J, Imoto I. Comparative genomic hybridization (CGH)-arrays pave the way for identification of novel cancer-related genes. Cancer Sci. 2004;95:559-63
139. Yu M, Ting DT, Stott SL, Wittner BS, Ozsolak F, Paul S. et al. RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis. Nature. 2012;487:510-3
140. Meng S, Tripathy D, Shete S, Ashfaq R, Haley B, Perkins S. et al. HER-2 gene amplification can be acquired as breast cancer progresses. Proc Natl Acad Sci U S A. 2004;101:9393-8
141. Miyamoto DT, Lee RJ, Stott SL, Ting DT, Wittner BS, Ulman M. et al. Androgen receptor signaling in circulating tumor cells as a marker of hormonally responsive prostate cancer. Cancer Discov. 2012;2:995-1003
142. Zhang S, Li L, Wang T, Bian L, Hu H, Xu C. et al. Real-time HER2 status detected on circulating tumor cells predicts different outcomes of anti-HER2 therapy in histologically HER2-positive metastatic breast cancer patients. BMC Cancer. 2016;16:526
143. Galletti G, Sung MS, Vahdat LT, Shah MA, Santana SM, Altavilla G. et al. Isolation of breast cancer and gastric cancer circulating tumor cells by use of an anti HER2-based microfluidic device. Lab on a Chip. 2013
144. Meng S, Tripathy D, Shete S, Ashfaq R, Saboorian H, Haley B. et al. uPAR and HER-2 gene status in individual breast cancer cells from blood and tissues. Proc Natl Acad Sci U S A. 2006;103:17361-5
145. Alunni-Fabbroni M, Sandri MT. Circulating tumour cells in clinical practice: Methods of detection and possible characterization. Methods. 2010;50:289-97
146. Kalinich M, Bhan I, Kwan TT, Miyamoto DT, Javaid S, LiCausi JA. et al. An RNA-based signature enables high specificity detection of circulating tumor cells in hepatocellular carcinoma. Proc Natl Acad Sci U S A. 2017;114:1123-8
147. Stott SL, Lee RJ, Nagrath S, Yu M, Miyamoto DT, Ulkus L. et al. Isolation and characterization of circulating tumor cells from patients with localized and metastatic prostate cancer. Sci Transl Med. 2010;2:25ra3
148. Ivanov I, Stojcic J, Stanimirovic A, Sargent E, Nam RK, Kelley SO. Chip-based nanostructured sensors enable accurate identification and classification of circulating tumor cells in prostate cancer patient blood samples. Anal Chem. 2013;85:398-403
149. Zhao R, Cai Z, Li S, Cheng Y, Gao H, Liu F. et al. Expression and clinical relevance of epithelial and mesenchymal markers in circulating tumor cells from colorectal cancer. Oncotarget. 2017;8:9293
150. Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT. et al. Circulating Breast Tumor Cells Exhibit Dynamic Changes in Epithelial and Mesenchymal Composition. Science. 2013;339:580-4
151. Kondo Y, Hayashi K, Kawakami K, Miwa Y, Hayashi H, Yamamoto M. KRAS mutation analysis of single circulating tumor cells from patients with metastatic colorectal cancer. BMC Cancer. 2017;17:311
152. Shao Diane D, Xue W, Krall Elsa B, Bhutkar A, Piccioni F, Wang X. et al. KRAS and YAP1 Converge to Regulate EMT and Tumor Survival. Cell. 2014;158:171-84
153. Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV. et al. Detection of Mutations in EGFR in Circulating Lung-Cancer Cells. New England Journal of Medicine. 2008;359:366-77
154. Ashby J, Flack K, Jimenez LA, Duan Y, Khatib A-K, Somlo G. et al. Distribution Profiling of Circulating MicroRNAs in Serum. Analytical Chemistry. 2014;86:9343-9
155. Di Meo A, Bartlett J, Cheng Y, Pasic MD, Yousef GM. Liquid biopsy: a step forward towards precision medicine in urologic malignancies. Mol Cancer. 2017;16:80
156. Melo S, Sugimoto H, O'Connell J, Kato N, Villanueva A, Vidal A. et al. Cancer Exosomes Perform Cell-Independent MicroRNA Biogenesis and Promote Tumorigenesis. Cancer Cell. 2014;26:707-21
157. Dawson S-J, Tsui DWY, Murtaza M, Biggs H, Rueda OM, Chin S-F. et al. Analysis of Circulating Tumor DNA to Monitor Metastatic Breast Cancer. New England Journal of Medicine. 2013;368:1199-209
158. Alix-Panabières C, Schwarzenbach H, Pantel K. Circulating Tumor Cells and Circulating Tumor DNA. Annual Review of Medicine. 2012;63:199-215
159. Wen CY, Wu LL, Zhang ZL, Liu YL, Wei SZ, Hu J. et al. Quick-Response Magnetic Nanospheres for Rapid, Efficient Capture and Sensitive Detection of Circulating Tumor Cells. Acs Nano. 2014;8:941
160. Chen Z, Hong G, Wang H, Welsher K, Tabakman SM, Sherlock SP. et al. Graphite-coated magnetic nanoparticle microarray for few-cells enrichment and detection. Acs Nano. 2012;6:1094
161. Wu CH, Huang YY, Chen P, Hoshino K, Liu H, Frenkel EP. et al. Versatile immunomagnetic nanocarrier platform for capturing cancer cells. Acs Nano. 2013;7:8816-23
162. Li W, Reategui E, Park MH, Castleberry S, Deng JZ, Hsu B. et al. Biodegradable nano-films for capture and non-invasive release of circulating tumor cells. Biomaterials. 2015;65:93-102
163. Talasaz AH, Powell AA, Huber DE, Berbee JG, Roh KH, Yu W. et al. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3970
164. Lohr JG, Adalsteinsson VA, Cibulskis K, Choudhury AD, Rosenberg M, Cruzgordillo P. et al. Whole exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nature Biotechnology. 2014;32:479-84
165. Huang YY, Hoshino K, Chen P, Wu CH, Lane N, Huebschman M. et al. Immunomagnetic nanoscreening of circulating tumor cells with a motion controlled microfluidic system. Biomed Microdevices. 2013;15:673-81
166. Kim W, Ng JK, Kunitake ME, Conklin BR, Yang P. Interfacing Silicon Nanowires with Mammalian Cells. Journal of the American Chemical Society. 2007;129:7228-9
167. Chua KN, Chai C, Lee PC, Tang YN, Ramakrishna S, Leong KW. et al. Surface-aminated electrospun nanofibers enhance adhesion and expansion of human umbilical cord blood hematopoietic stem/progenitor cells. Biomaterials. 2006;27:6043-51
Author contact
![]() Corresponding authors: shichenedu.cn; lianrongedu.cn
Corresponding authors: shichenedu.cn; lianrongedu.cn

 Global reach, higher impact
Global reach, higher impact